UniProt ID: P22303-1 (32-578) acetylcholinesterase
UniProt ID: P22303-1 (32-578) acetylcholinesterase
|
|
|
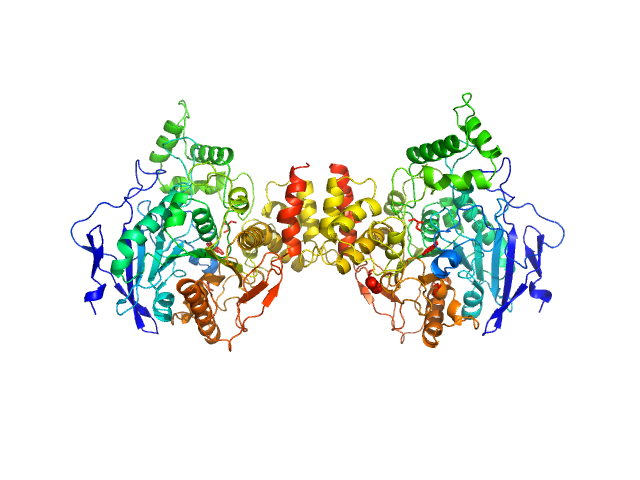
|
| Sample: |
Acetylcholinesterase dimer, 120 kDa Homo sapiens protein
Acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|