|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (wild-type, residues 708-801, i.e. ADAR1-dsRBD3) dimer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (interface mutant, residues 708-801, i.e. ADAR1-dsRBD3 interface mutant) monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Chimeric construct between the third dsRBD of human ADAR1 and the second dsRBD of Xenopus Xlrbpa (chimeric construct, residues 708-801, i.e. Chimeric ADAR1-ds3/Xlrbpa-ds2) monomer, 13 kDa Homo sapiens / … protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
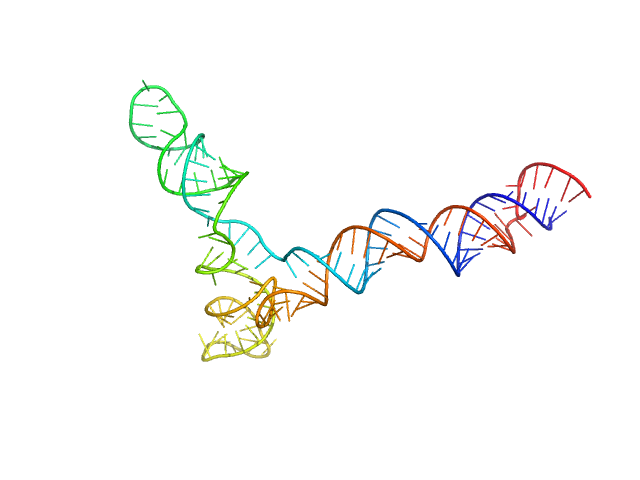
| Sample: |
Full stem-loop 5 of SARS-CoV-2 5'genomic end monomer, 48 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.8 |
nm |
|
|
|
|
|
|

|
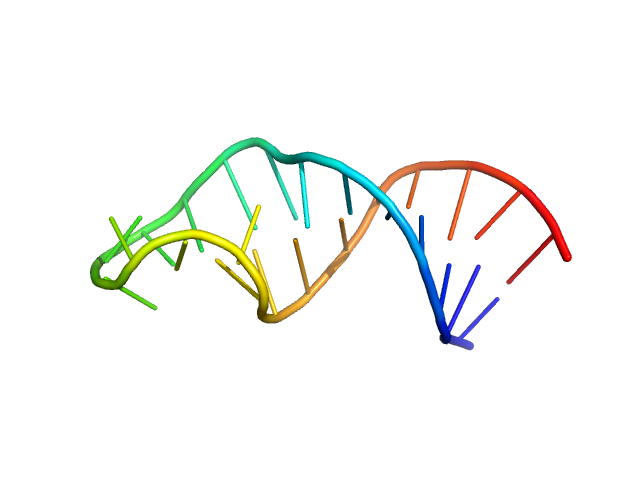
| Sample: |
Sub-element stem-loop 5a from SARS-CoV-2 5'genomic end monomer, 11 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Sub-element stem-loop 5b from SARS-CoV-2 5'genomic end monomer, 8 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Feb 3
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
|
|
|
|
|
|
|
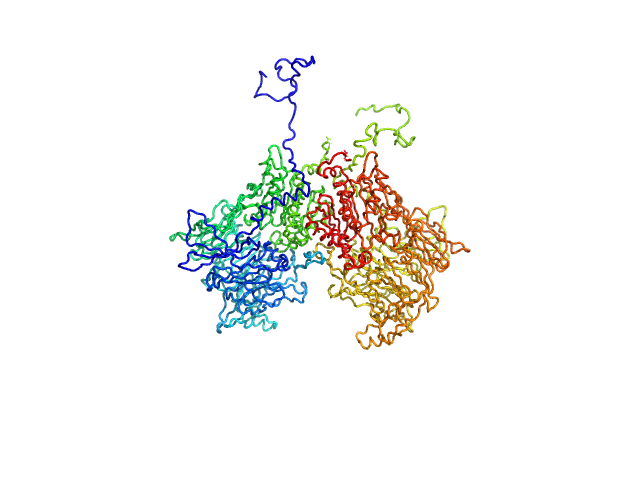
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
271 |
nm3 |
|
|
|
|
|
|
|
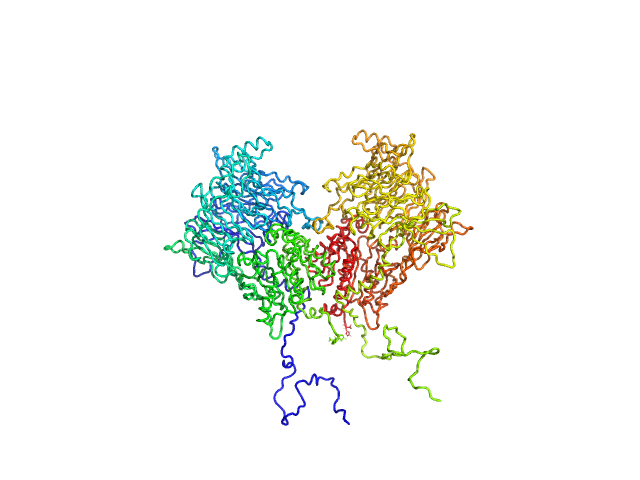
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
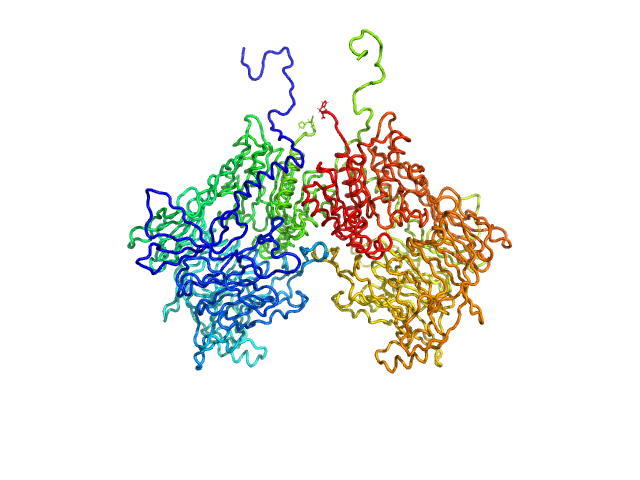
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
|
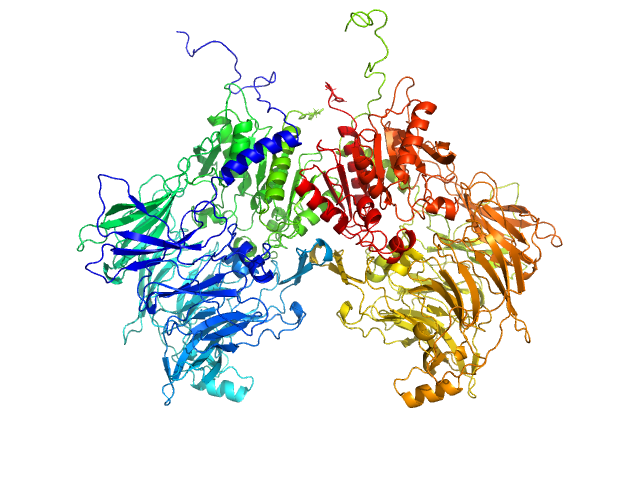
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
274 |
nm3 |
|
|