|
|
|
|
|
| Sample: |
Elastin-like polypeptide monomer, 13 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline containing 6 M urea, 0.3 M KCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 2
|
A genetically encoded anomalous SAXS ruler to probe the dimensions of intrinsically disordered proteins.
Proc Natl Acad Sci U S A 121(50):e2415220121 (2024)
Yu M, Gruzinov AY, Ruan H, Scheidt T, Chowdhury A, Giofrè S, Mohammed ASA, Caria J, Sauter PF, Svergun DI, Lemke EA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Elastin-like polypeptide labeled with unnatural amino acid diBrK monomer, 13 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline containing 6 M urea, 0.3 M KCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 2
|
A genetically encoded anomalous SAXS ruler to probe the dimensions of intrinsically disordered proteins.
Proc Natl Acad Sci U S A 121(50):e2415220121 (2024)
Yu M, Gruzinov AY, Ruan H, Scheidt T, Chowdhury A, Giofrè S, Mohammed ASA, Caria J, Sauter PF, Svergun DI, Lemke EA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
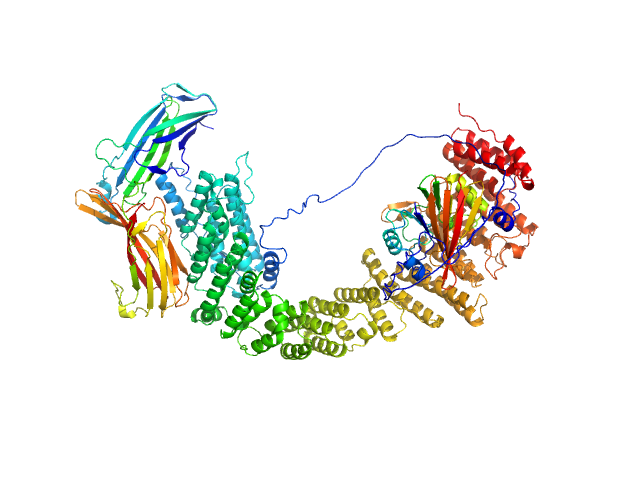
| Sample: |
VPS35 endosomal protein-sorting factor-like monomer, 110 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 29 monomer, 21 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Apr 28
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
|
|
|
|
|
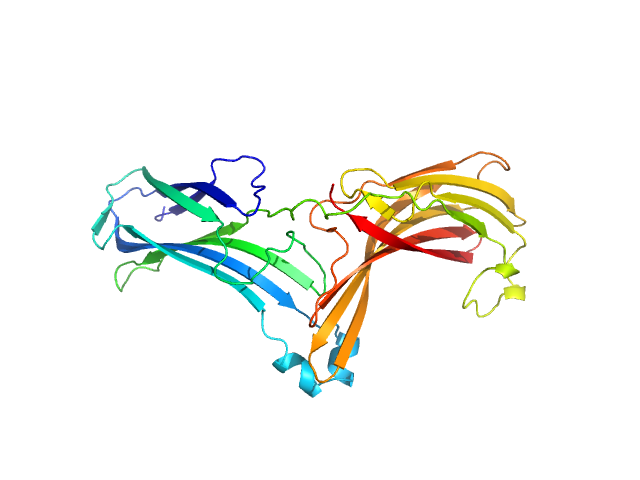
| Sample: |
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 22
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
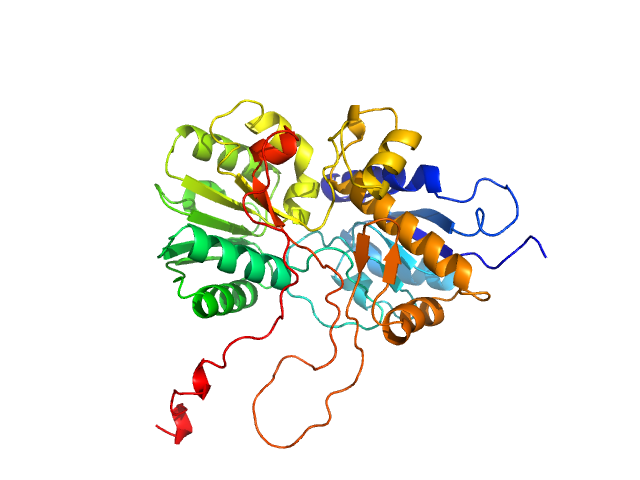
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
MRNA-capping enzyme catalytic subunit monomer, 35 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Adenylate cyclase monomer, 43 kDa Trypanosoma brucei protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Biophysical analysis of the membrane-proximal Venus Flytrap domain of ESAG4 receptor-like adenylate cyclase from Trypanosoma brucei.
Mol Biochem Parasitol 260:111653 (2024)
Alves DO, Geens R, da Silva Arruda HR, Jennen L, Corthaut S, Wuyts E, de Andrade GC, Prosdocimi F, Cordeiro Y, Pires JR, Vieira LR, de Oliveira GAP, Sterckx YG, Salmon D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 688-817, i.e. dsRBD3-long) dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 708-801, i.e. dsRBD3-mid) dimer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
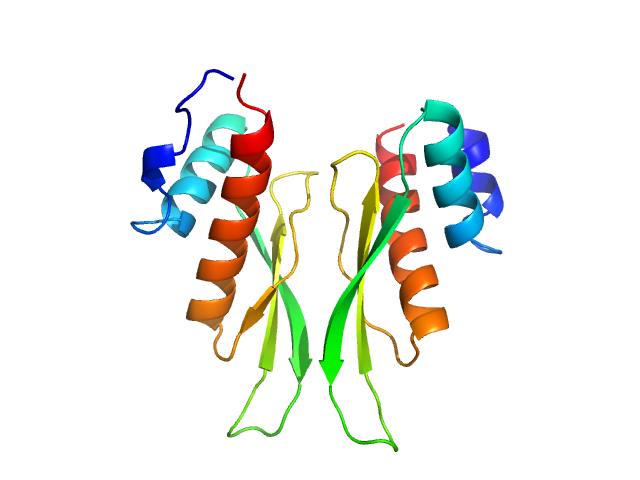
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 716-797, i.e. dsRBD3-short) dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|