|
|
|
|
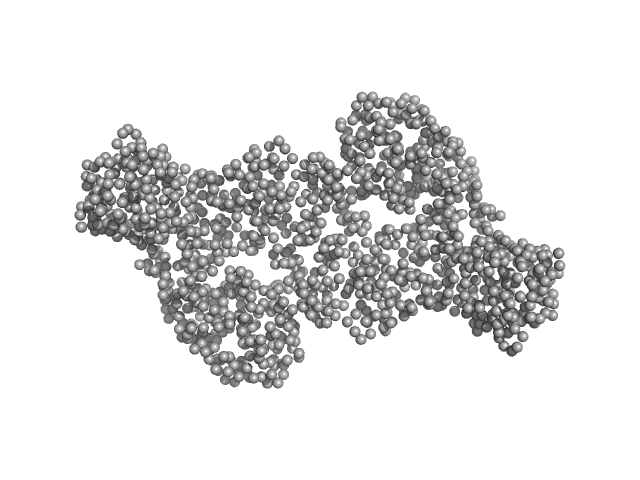
|
| Sample: |
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
DNA methyltransferase 3 beta (413-853) dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Nov 2
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
191 |
nm3 |
|
|
|
|
|
|
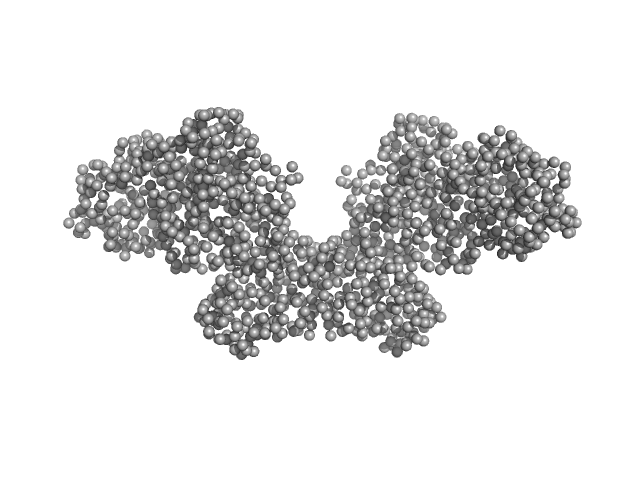
|
| Sample: |
Histone H3.3 dimer, 8 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
DNA methyltransferase 3 beta (413-853) dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Nov 2
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
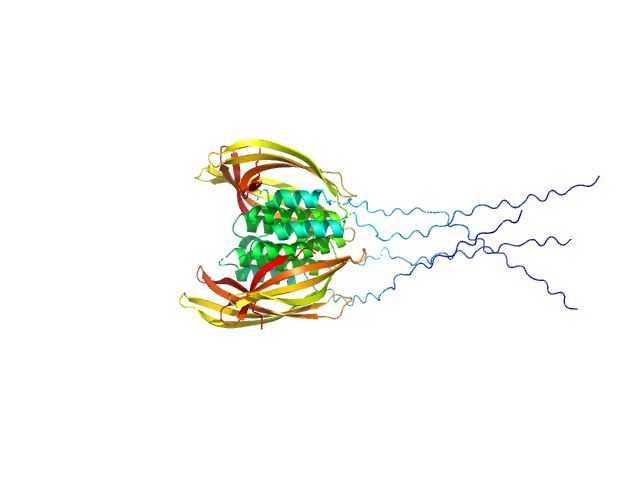
|
| Sample: |
Probable conserved Mce associated membrane protein tetramer, 59 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM Tris, 500mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 10
|
Insight into the structure and interactions of the M. tuberculosis
Mce-associated membrane proteins Mam1A-1D
(2025)
Hynönen M, Perumal P, Hynönen N, Doutch J, Ma K, Venkatesan R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
90.0 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Probable conserved Mce associated membrane protein tetramer, 59 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, D-C12E9, pH: 8.5 |
| Experiment: |
SANS
data collected at SANS2D, ISIS Neutron and Muon Source on 2019 Dec 10
|
Insight into the structure and interactions of the M. tuberculosis
Mce-associated membrane proteins Mam1A-1D
(2025)
Hynönen M, Perumal P, Hynönen N, Doutch J, Ma K, Venkatesan R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Jun 2
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphorylated Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Jun 2
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
3.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 500 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Nov 19
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphorylated Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 500 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Nov 19
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Heterochromatin protein HP1α, delta CSD mutant monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 27
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphorylated Heterochromatin protein HP1α, delta CSD mutant monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 27
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|