|
|
|
|
|
| Sample: |
Human immunoglobulin SAP1.3 - kappa chain dimer, 48 kDa Homo sapiens protein
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain dimer, 99 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain C226S dimer, 99 kDa Homo sapiens protein
Human immunoglobulin SAP1.3 - kappa chain C214S dimer, 48 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human immunoglobulin SAP1.3 - kappa chain C214S dimer, 48 kDa Homo sapiens protein
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain C227S dimer, 99 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human immunoglobulin SAP1.3 - kappa chain dimer, 48 kDa Homo sapiens protein
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain C226S/C227S dimer, 99 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|
|
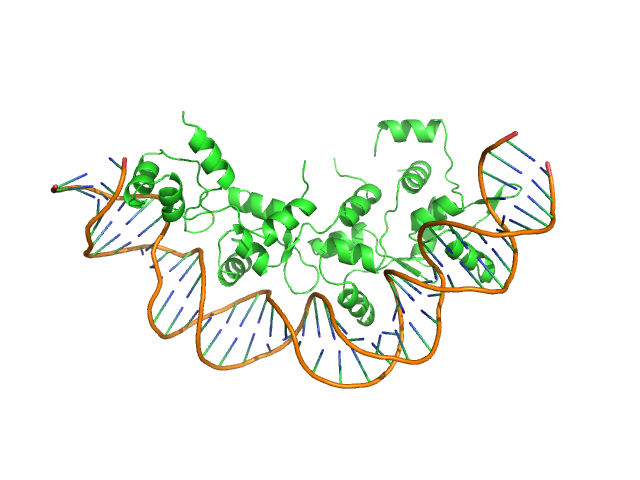
| Sample: |
Recombination directionality factor RdfS tetramer, 52 kDa Mesorhizobium japonicum R7A protein
AttP_8 40-mer DNA dimer, 25 kDa DNA
|
| Buffer: |
150 mM Tris-HCl, 300 mM NaCl, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 19
|
Structural basis for control of integrative and conjugative element excision and transfer by the oligomeric winged helix–turn–helix protein RdfS
Nucleic Acids Research 53(6) (2025)
Verdonk C, Agostino M, Eto K, Hall D, Bond C, Ramsay J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDVV6_fit1_model1.png)
|
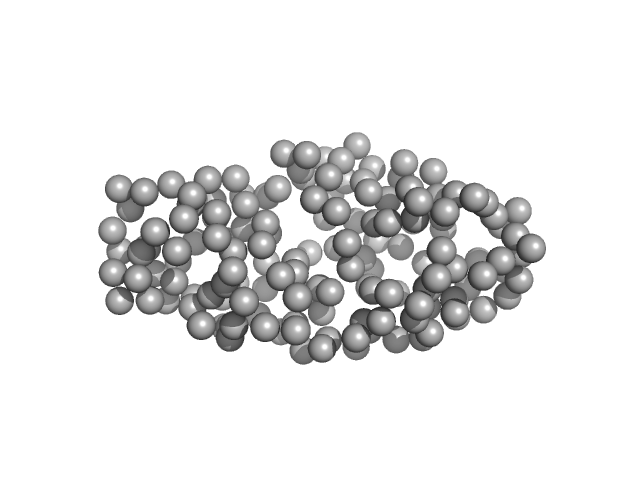
| Sample: |
Drebrin monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
17 mM NaH2PO4, 3 mM Na2HPO4, 50 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 7
|
Dynamic Interchange of Local Residue-Residue Interactions in the Largely Extended Single Alpha-Helix in Drebrin
Biochemical Journal (2025)
Varga S, Péterfia B, Dudola D, Farkas V, Jeffries C, Permi P, Gáspári Z
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Apr 20
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
20464 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
271263 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Histone H3.3 dimer, 8 kDa Homo sapiens protein
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Histone H3.3 monomer, 4 kDa Homo sa protein
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Sep 17
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
19 |
nm3 |
|
|