|
|
|
|
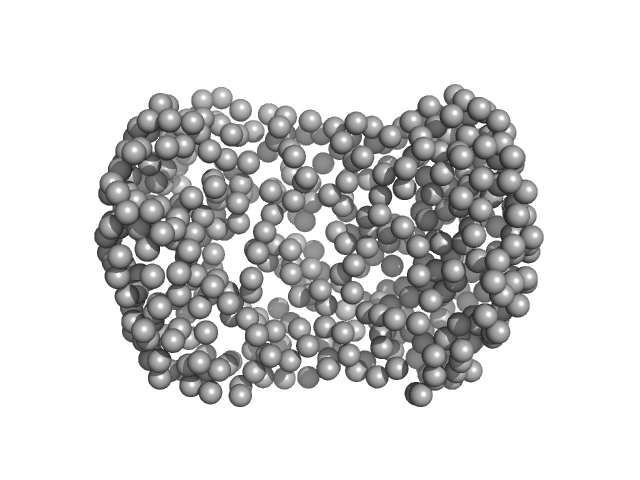
|
| Sample: |
Auxin response factor monomer, 46 kDa Marchantia polymorpha protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
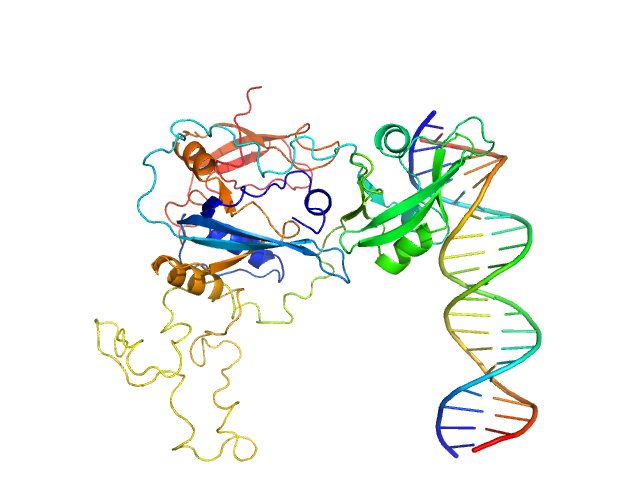
|
| Sample: |
Auxin response factor monomer, 46 kDa Marchantia polymorpha protein
High Affinity ARF binding sequence inverted repeat with 6 nucleotide spacing dimer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Auxin response factor 1 dimer, 81 kDa Arabidopsis thaliana protein
High Affinity ARF binding sequence inverted repeat with 7 nucleotide spacing dimer, 13 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W (artificial PNT3 variant - low-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.8 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W (artificial PNT3 variant - high-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 5
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-lactoglobulin, 20 kDa Bos taurus protein
|
| Buffer: |
20 mM MOPS, 100 mM NaCl, 1 mM TCEP, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 21
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-lactoglobulin, 20 kDa Bos taurus protein
|
| Buffer: |
20 mM MOPS, 100 mM NaCl, 1 mM TCEP, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 21
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-lactoglobulin, 20 kDa Bos taurus protein
|
| Buffer: |
20 mM MOPS, 100 mM NaCl, 1 mM TCEP, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 21
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-lactoglobulin, 20 kDa Bos taurus protein
|
| Buffer: |
20 mM MOPS, 100 mM NaCl, 1 mM TCEP, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 21
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
40 |
nm3 |
|
|