|
|
|
|
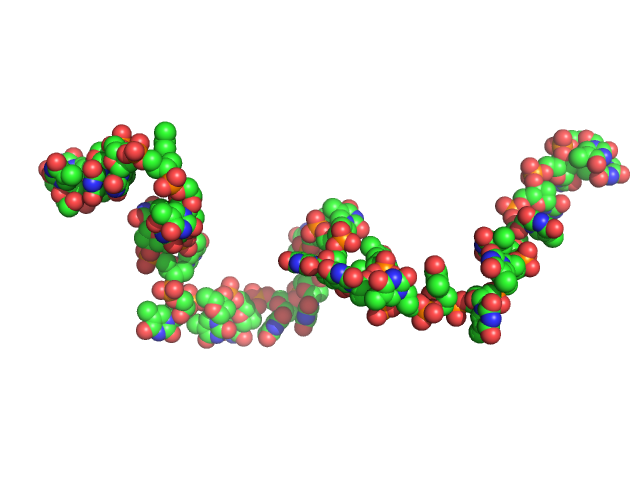
|
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 200mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
94673 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, on 2015 Feb 1
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
70673 |
nm3 |
|
|
|
|
|
|
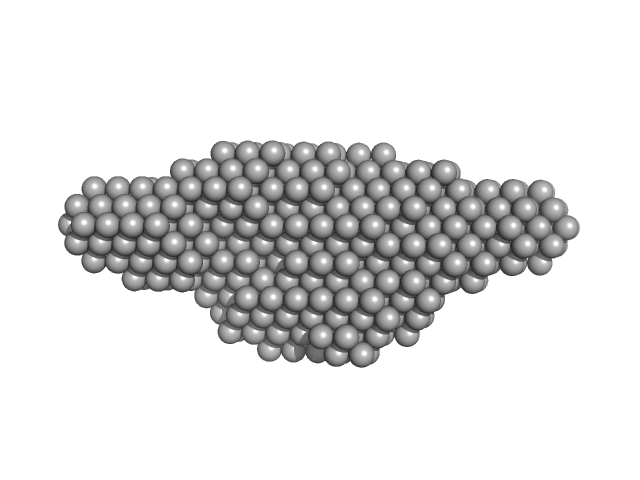
|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) dimer, 166 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) N domain monomer, 26 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) N and M domains monomer, 62 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
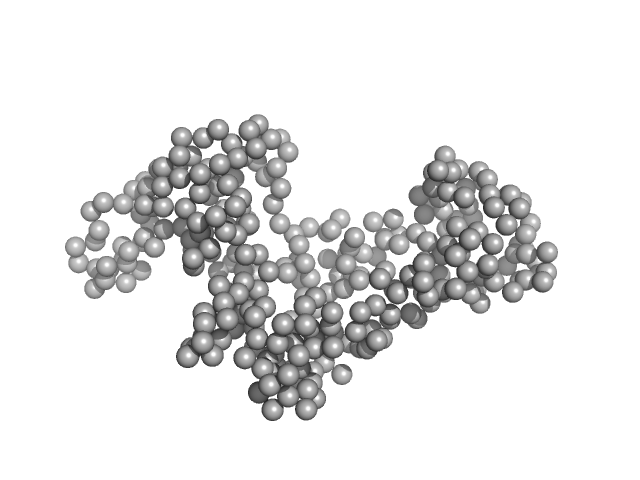
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 31 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 20
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
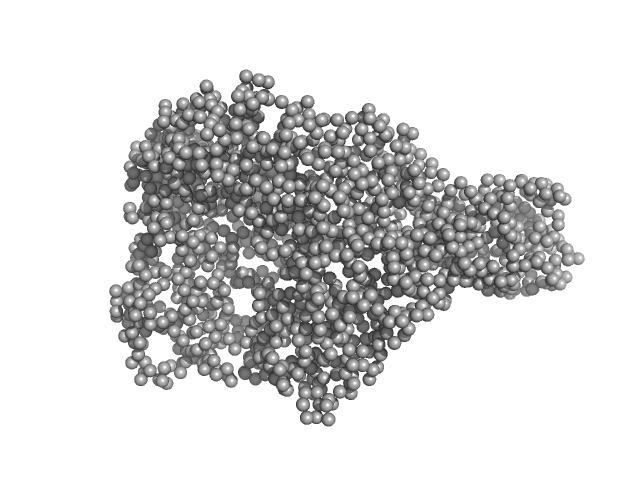
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 135 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
280 |
nm3 |
|
|