|
|
|
|
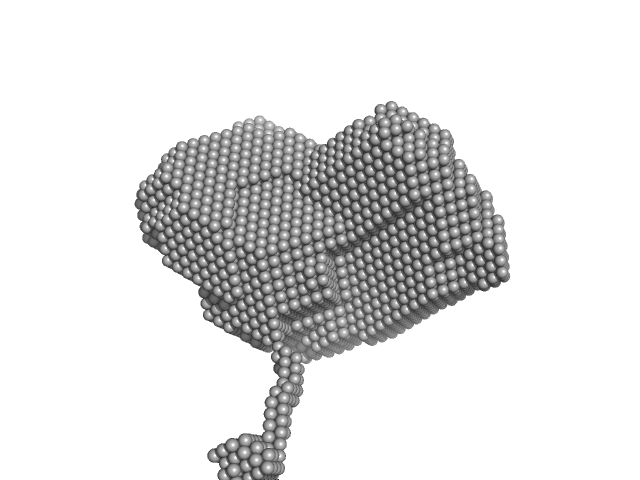
|
| Sample: |
Group IIC Intron Domain 1 monomer, 89 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 7
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|
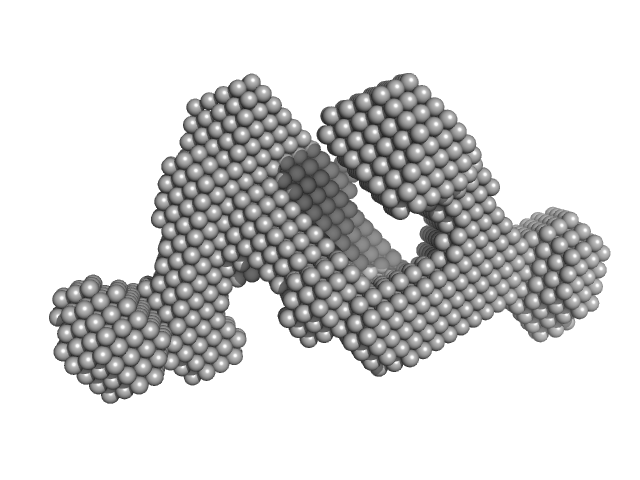
|
| Sample: |
Group IIC Intron Domain 1, 2 & 3 monomer, 103 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 2
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
195 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Group IIC Intron Domain 1 & 2 monomer, 96 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 2
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
179 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Group IIC intron Domain 1, 2, 3 & 4 monomer, 125 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 2
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
247 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Estrogen receptor alpha dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Nov 9
|
A ternary switch model governing ERα ligand binding domain conformation.
Nat Commun 16(1):10363 (2025)
McDougal DP, Pederick JL, Novick SJ, Jovcevski B, Warrender AK, Pascal BD, Griffin PR, Bruning JB
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Estrogen receptor alpha dimer, 57 kDa Melanotaenia fluviatilis protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Nov 9
|
A ternary switch model governing ERα ligand binding domain conformation.
Nat Commun 16(1):10363 (2025)
McDougal DP, Pederick JL, Novick SJ, Jovcevski B, Warrender AK, Pascal BD, Griffin PR, Bruning JB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Kinesin-associated protein 3 monomer, 81 kDa Mus musculus protein
Kinesin-like protein KIF3B dimer, 64 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 14
|
Kinesin protein 3 B/B and kinesin associated protein 3 (KAP)
Kento Yonezawa
|
| RgGuinier |
5.7 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
385 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bovine Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Jul 9
|
RnaseA
Anthony Giannetti
|
| RgGuinier |
1.6 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hen egg white lysozyme monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Jul 9
|
Lysozyme pH 7.4 with models
Anthony Giannetti
|
| RgGuinier |
1.6 |
nm |
| Dmax |
59.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
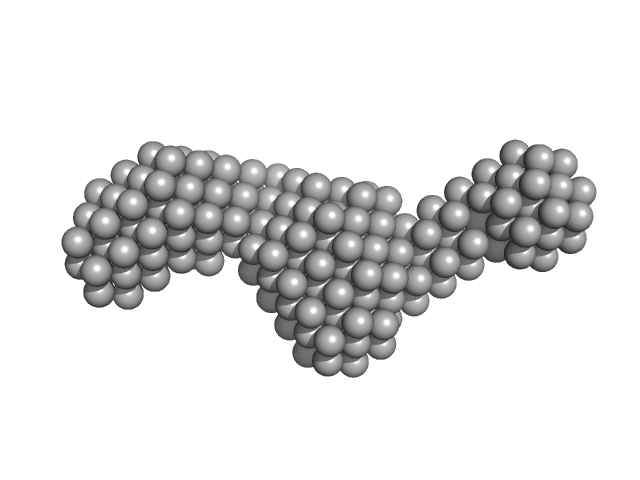
|
| Sample: |
Histatin-3 monomer, 4 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Jul 9
|
Histatin 3
Anthony Giannetti
|
| RgGuinier |
1.5 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
7 |
nm3 |
|
|