|
|
|
|

|
| Sample: |
Mouse ProNGF - pro-form of the NGF (nerve growth factor) dimer, 100 kDa Mus musculus protein
|
| Buffer: |
50 mM Na-phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 18
|
Structural and functional properties of mouse proNGF.
Biochem Soc Trans 34(Pt 4):605-6 (2006)
Paoletti F, Konarev PV, Covaceuszach S, Schwarz E, Cattaneo A, Lamba D, Svergun DI
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polypyrimidine tract-binding protein 2 monomer, 57 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 250 mM NaCl, 2 mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 11
|
Structure and RNA interactions of the N-terminal RRM domains of PTB.
Structure 12(9):1631-43 (2004)
Simpson PJ, Monie TP, Szendröi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthews S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Procollagen C-endopeptidase enhancer 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Jul 9
|
Low Resolution Structure Determination Shows Procollagen C-Proteinase Enhancer to be an Elongated Multidomain Glycoprotein
Journal of Biological Chemistry 278(9):7199-7205 (2003)
Bernocco S, Steiglitz B, Svergun D, Petoukhov M, Ruggiero F, Ricard-Blum S, Ebel C, Geourjon C, Deléage G, Font B, Eichenberger D, Greenspan D, Hulmes D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
264 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.3 |
nm |
| Dmax |
18.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.5 |
nm |
|
|
|
|
|
|
|
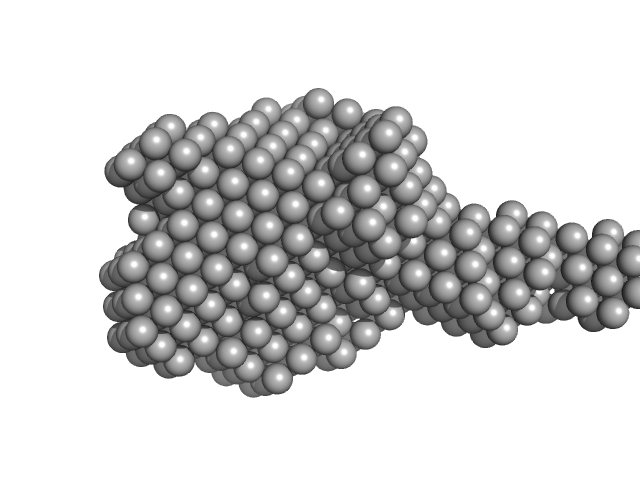
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 processively polymerizes length-controlled matriglycan on prodystroglycan.
Nat Commun 16(1):9028 (2025)
Joseph S, Schnicker NJ, Spellmon N, Xu Z, Yan R, Yu Z, Davulcu O, Yang T, Hopkins J, Anderson ME, Venzke D, Campbell KP
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
231 |
nm3 |
|
|