|
|
|
|
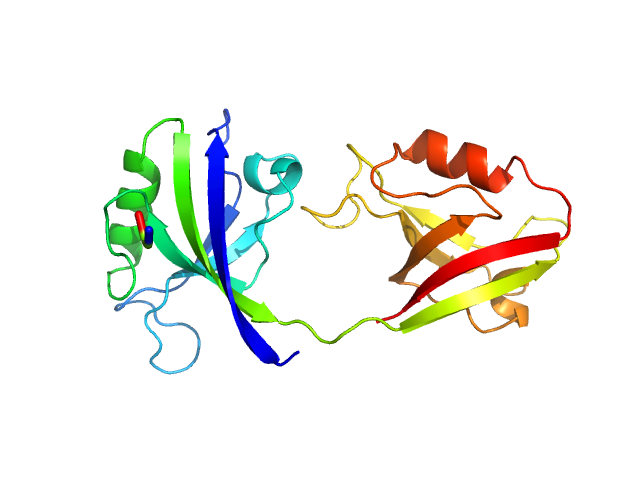
|
| Sample: |
PDZ1-2 fragment of PSD-95/Disks large homolog 4 monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM TRIS/HCl, 150 mM NaCl + 10 mM GSH, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jan 26
|
The dual PDZ domain from Postsynaptic density protein 95 forms a scaffold with peptide ligand
Biophysical Journal (2020)
Rodzli N, Lockhart-Cairns M, Levy C, Chipperfield J, Bird L, Baldock C, Prince S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
Condensin complex subunit 2 monomer, 17 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
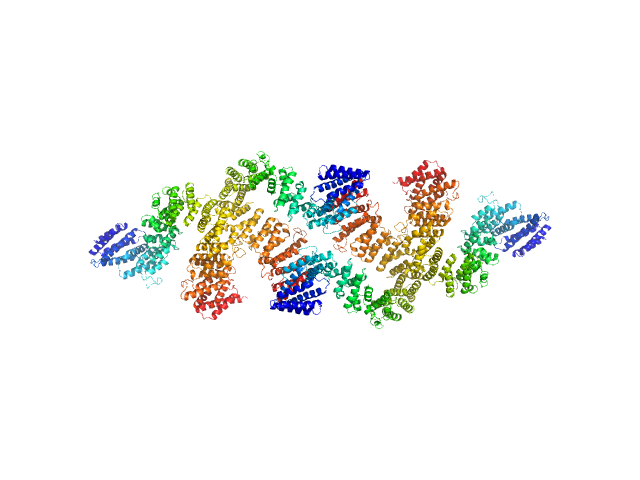
|
| Sample: |
Condensin complex subunit 3-like protein tetramer, 433 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
8.3 |
nm |
| Dmax |
32.7 |
nm |
| VolumePorod |
1080 |
nm3 |
|
|
|
|
|
|
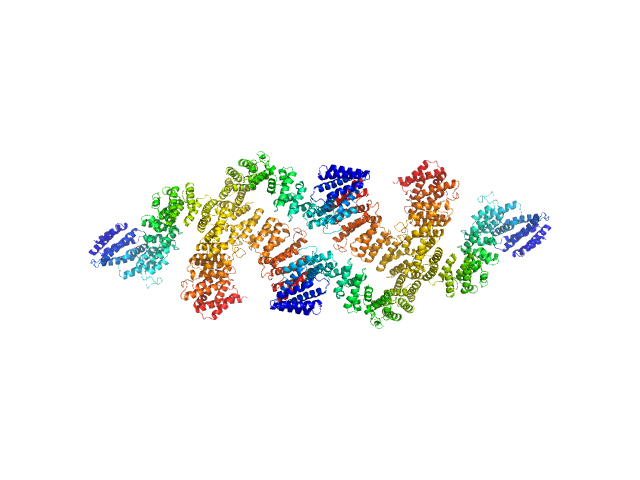
|
| Sample: |
Condensin complex subunit 3-like protein tetramer, 433 kDa Chaetomium thermophilum protein
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
6.8 |
nm |
| Dmax |
29.8 |
nm |
| VolumePorod |
720 |
nm3 |
|
|
|
|
|
|
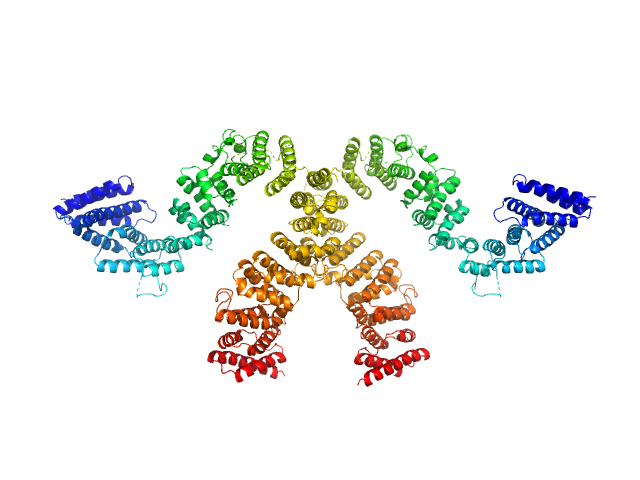
|
| Sample: |
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
463 |
nm3 |
|
|
|
|
|
|
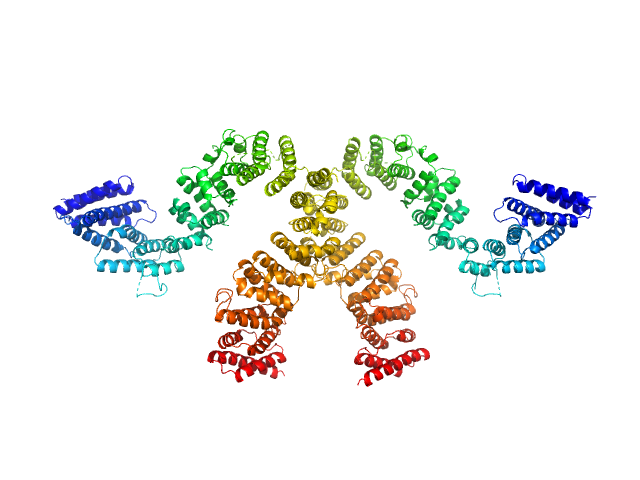
|
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
5.3 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
|
|
|
|
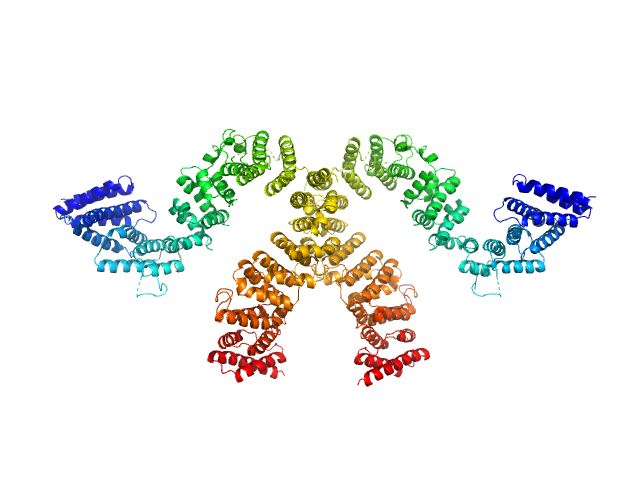
|
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
301 |
nm3 |
|
|
|
|
|
|
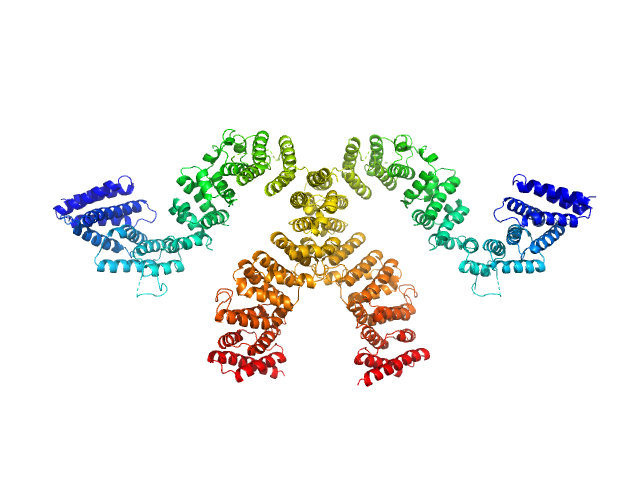
|
| Sample: |
Condensin complex subunit 3-like protein monomer, 108 kDa Chaetomium thermophilum protein
Condensin complex subunit 3-like protein dimer, 217 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 20
|
Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1.
J Biol Chem 294(37):13822-13829 (2019)
Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
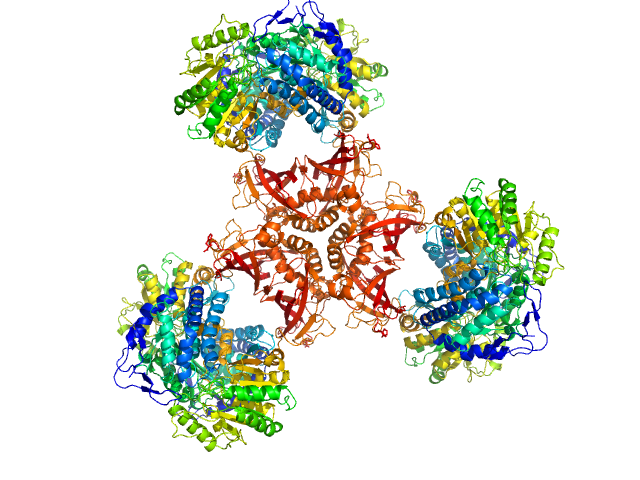
|
| Sample: |
Bifunctional protein PaaZ hexamer, 438 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Feb 24
|
Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun 10(1):4127 (2019)
Sathyanarayanan N, Cannone G, Gakhar L, Katagihallimath N, Sowdhamini R, Ramaswamy S, Vinothkumar KR
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
636 |
nm3 |
|
|