|
|
|
|
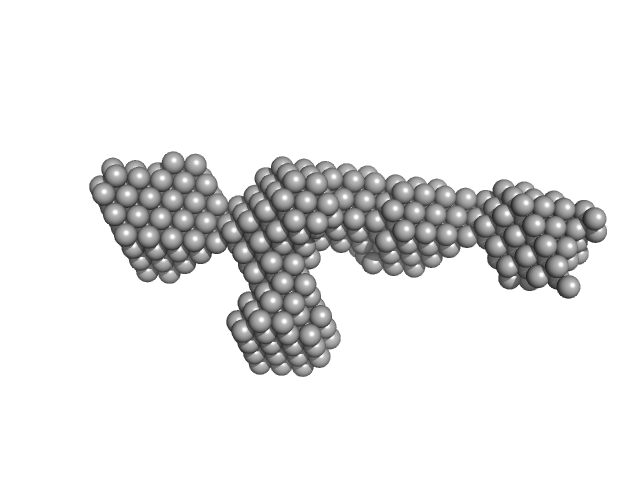
|
| Sample: |
S-layer protein monomer, 116 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
Water with Ca2+, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Analysis of self-assembly of S-layer protein slp-B53 from Lysinibacillus sphaericus.
Eur Biophys J 46(1):77-89 (2017)
Liu J, Falke S, Drobot B, Oberthuer D, Kikhney A, Guenther T, Fahmy K, Svergun D, Betzel C, Raff J
|
| RgGuinier |
6.4 |
nm |
| Dmax |
28.1 |
nm |
| VolumePorod |
609 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
S-layer protein monomer, 116 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
Water with Mg2+, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Analysis of self-assembly of S-layer protein slp-B53 from Lysinibacillus sphaericus.
Eur Biophys J 46(1):77-89 (2017)
Liu J, Falke S, Drobot B, Oberthuer D, Kikhney A, Guenther T, Fahmy K, Svergun D, Betzel C, Raff J
|
| RgGuinier |
6.6 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
597 |
nm3 |
|
|
|
|
|
|
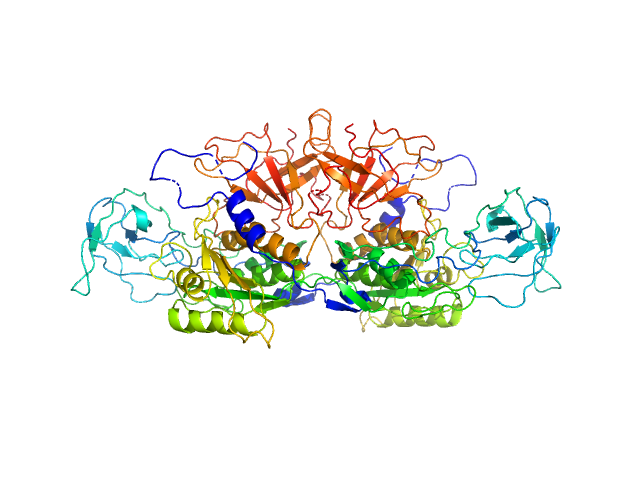
|
| Sample: |
Molybdopterin biosynthesis protein CNX1 dimer, 99 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Hepes/KOH, 150 mM NaCl, 1 mM EDTA, 2 % (v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2015 Feb 5
|
Dimerization of the plant molybdenum insertase Cnx1E is required for synthesis of the molybdenum cofactor.
Biochem J 474(1):163-178 (2017)
Krausze J, Probst C, Curth U, Reichelt J, Saha S, Schafflick D, Heinz DW, Mendel RR, Kruse T
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
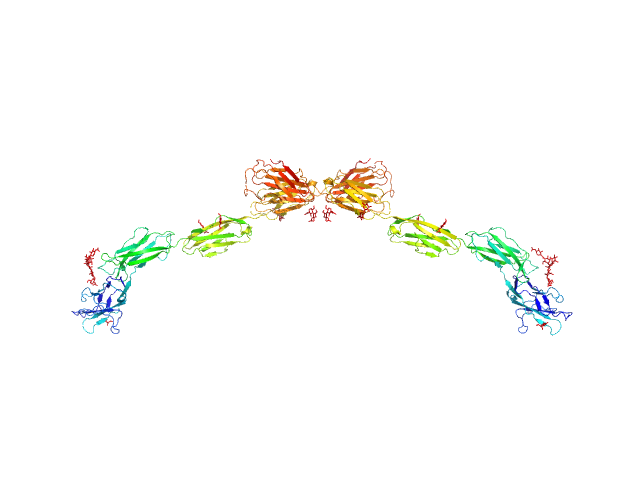
|
| Sample: |
Myelin-associated glycoprotein Ig domains 1-5 dimer, 108 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.8 |
nm |
| Dmax |
23.8 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
|
|
|
|
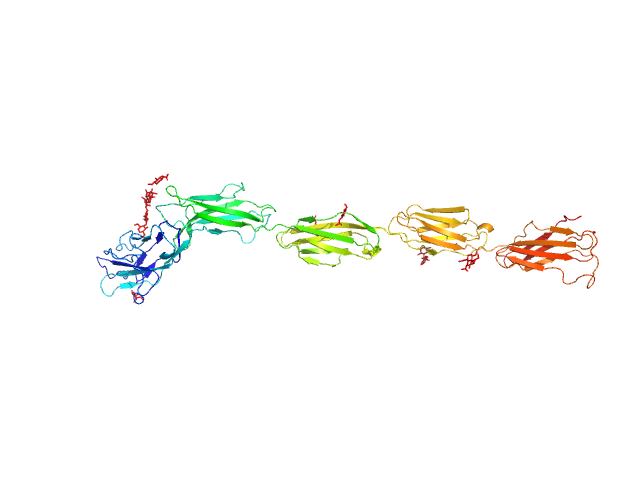
|
| Sample: |
Myelin-associated glycoprotein (20-508; I473E mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
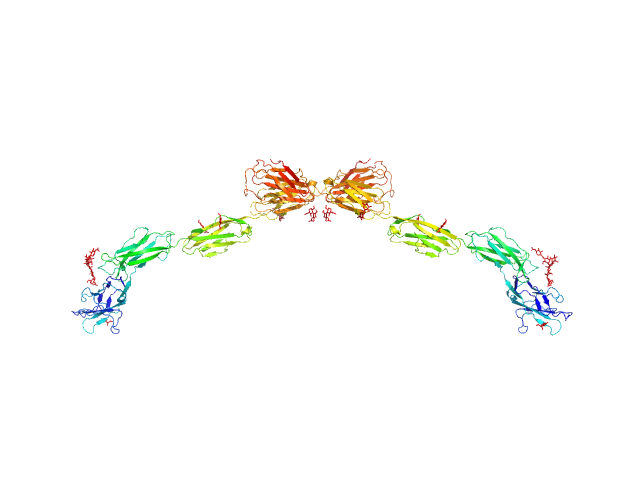
| Sample: |
Myelin-associated glycoprotein (20-508; N406Q mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
7.3 |
nm |
| Dmax |
25.6 |
nm |
| VolumePorod |
193 |
nm3 |
|
|
|
|
|
|
|
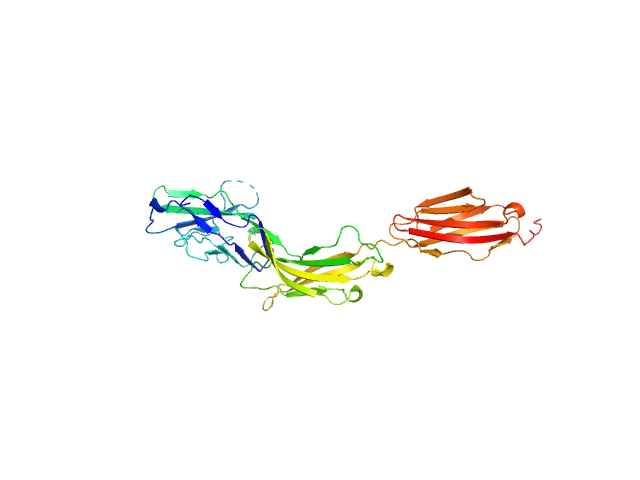
| Sample: |
Myelin-associated glycoprotein Ig domains 1-3 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
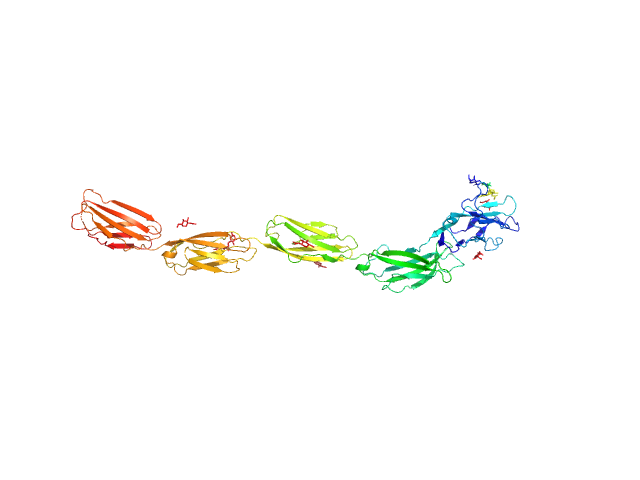
| Sample: |
Myelin-associated glycoprotein (20-508; I473E mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.0 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
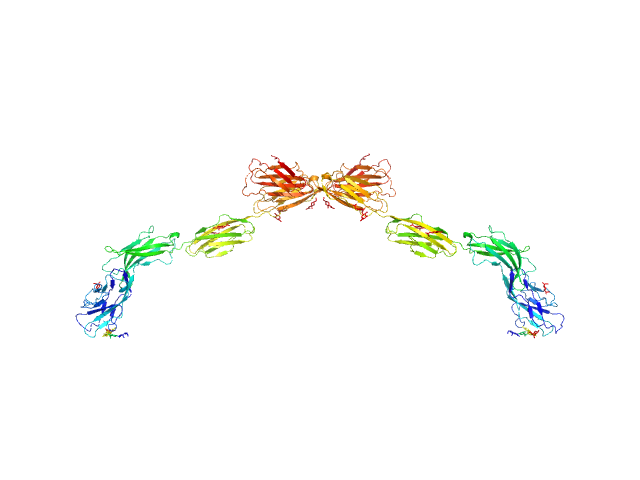
| Sample: |
Myelin-associated glycoprotein (20-508; N406Q mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jul 28
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
7.8 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
216 |
nm3 |
|
|
|
|
|
|
|
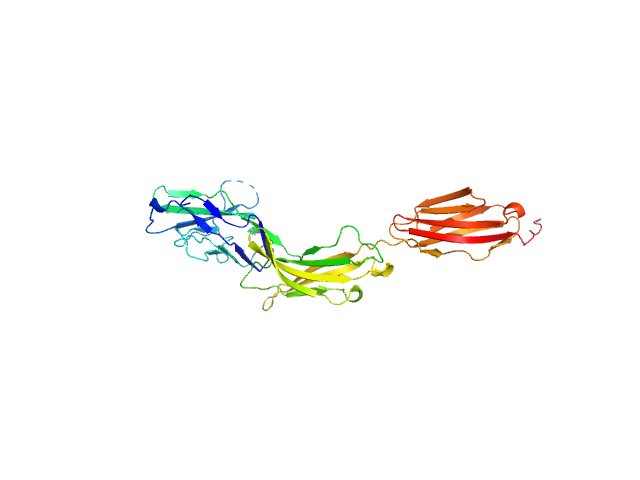
| Sample: |
Myelin-associated glycoprotein Ig domains 1-3 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
49 |
nm3 |
|
|