|
|
|
|
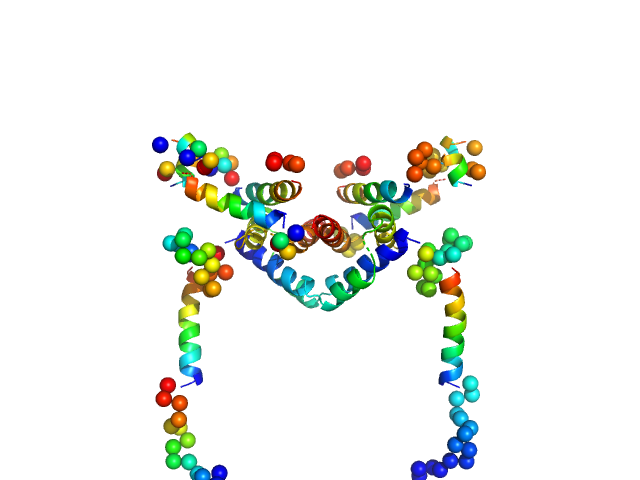
|
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|
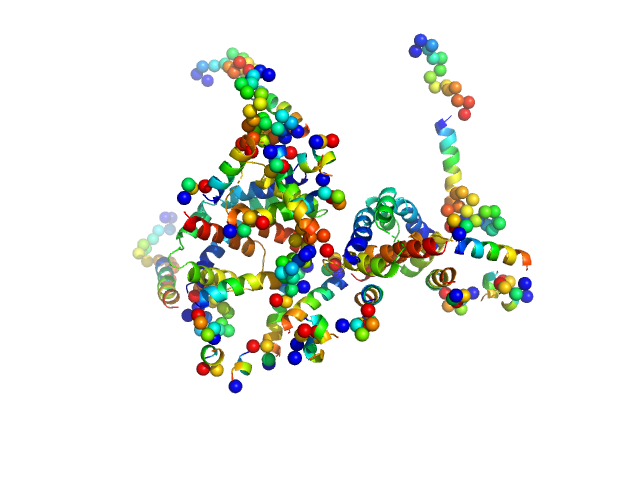
|
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) tetramer, 85 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
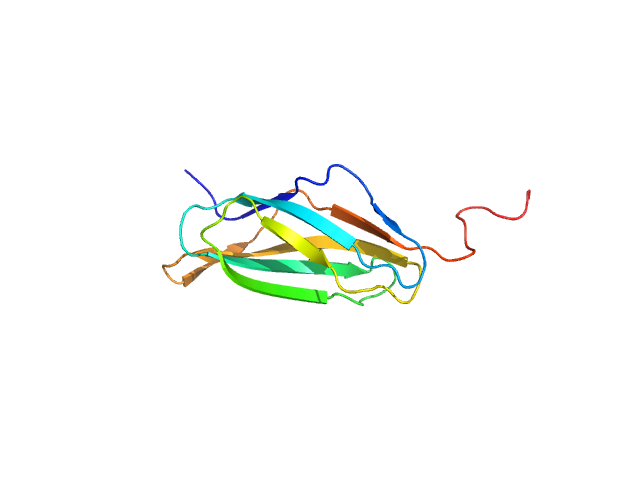
|
| Sample: |
Nucleoporin POM152 monomer, 12 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 10%(v/v) glycerol, 5mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Apr 12
|
Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure 25(3):434-445 (2017)
Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
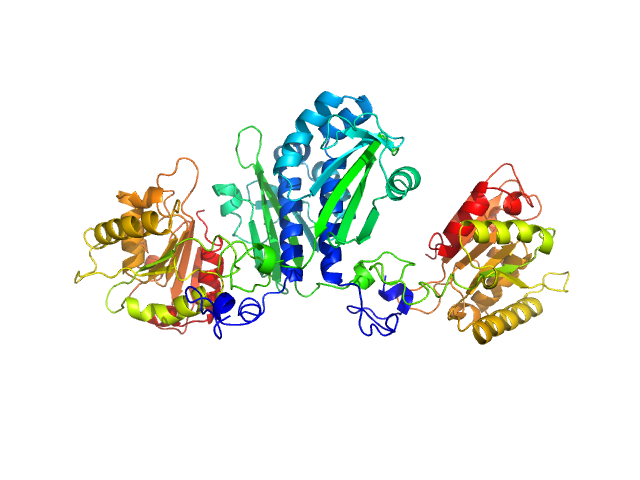
|
| Sample: |
Citrate-binding CitAP domain fused to lipase A of Bacillus subtilis BsLA dimer, 77 kDa protein
|
| Buffer: |
10 mM glycine buffer, 10 mM NaCl, 1 mM sodium citrate, pH: 10 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 6
|
A combination of mutational and computational scanning guides the design of an artificial ligand-binding controlled lipase.
Sci Rep 7:42592 (2017)
Kaschner M, Schillinger O, Fettweiss T, Nutschel C, Krause F, Fulton A, Strodel B, Stadler A, Jaeger KE, Krauss U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
|
|
|
|
|
|
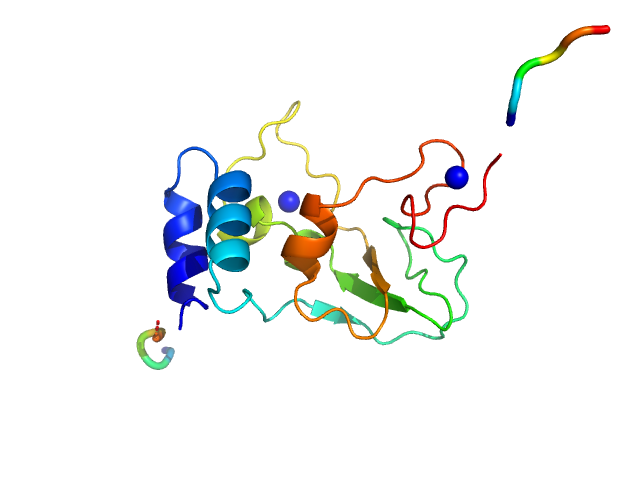
|
| Sample: |
HCoV-229E Non-structural protein 10 monomer, 15 kDa Human coronavirus 229E protein
|
| Buffer: |
25 mM HEPES 280 mM NaCl 2 mM DTT 500 µM ZnCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 25
|
Human alphacoronavirus non-structural protein Nsp10
Sven Falke,
Al Kikhney
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.8 |
nm |
|
|
|
|
|
|
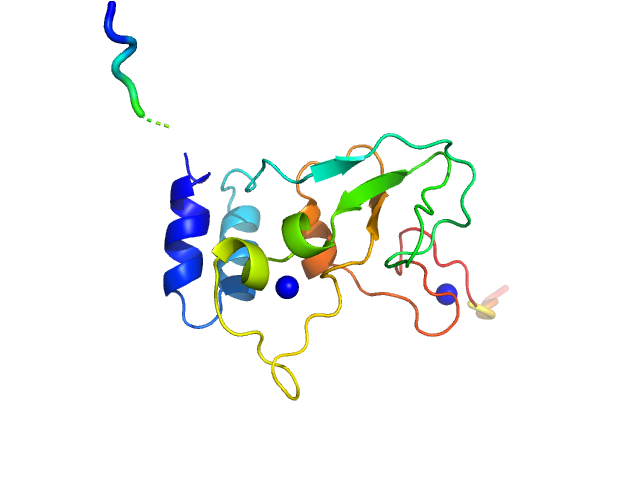
|
| Sample: |
HCoV-229E Non-structural protein 10 monomer, 15 kDa Human coronavirus 229E protein
|
| Buffer: |
25 mM HEPES 400 mM NaCl 1 mM EDTA 5% glycerol 40 mM NaH2PO4, pH: 7.9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2011 Sep 8
|
Human alphacoronavirus non-structural protein Nsp10
Sven Falke,
Al Kikhney
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.9 |
nm |
|
|
|
|
|
|
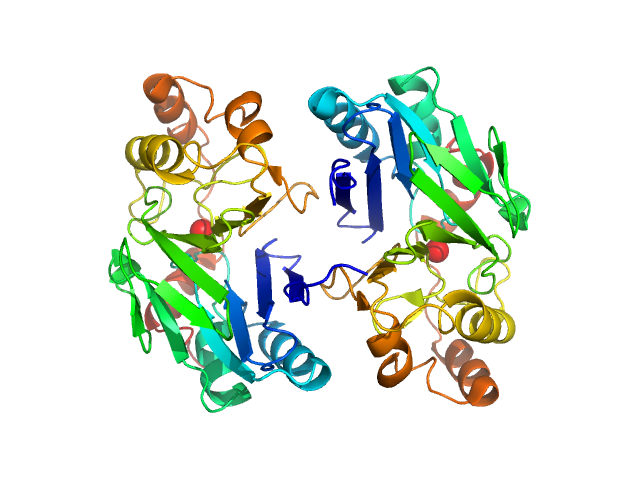
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 21
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
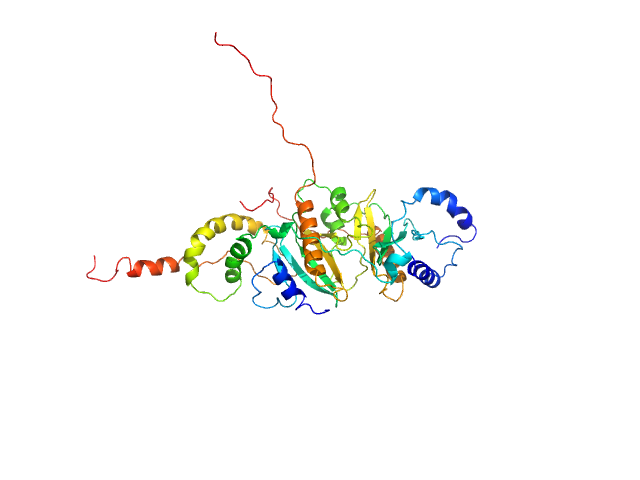
|
| Sample: |
SycH putative yopH targeting protein dimer, 32 kDa Yersinia pseudotuberculosis protein
Tyrosine-protein phosphatase YopH monomer, 14 kDa Yersinia pseudotuberculosis protein
|
| Buffer: |
50 mM HEPES 2mM TCEP, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Aug 1
|
Global Disordering in Stereo-Specific Protein Association
Biophysical Journal 112(3):33a (2017)
Gupta A, Reinartz I, Spilotros A, Jonna V, Hofer A, Svergun D, Schug A, Wolf-Watz M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-regulated outer membrane lipoprotein FrpD monomer, 27 kDa Neisseria meningitidis protein
|
| Buffer: |
10 mM Tris-HCl 150 mM NaCl 0.01% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Oct 19
|
Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep 7:40408 (2017)
Sviridova E, Rezacova P, Bondar A, Veverka V, Novak P, Schenk G, Svergun DI, Kuta Smatanova I, Bumba L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
S-layer protein monomer, 116 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Analysis of self-assembly of S-layer protein slp-B53 from Lysinibacillus sphaericus.
Eur Biophys J 46(1):77-89 (2017)
Liu J, Falke S, Drobot B, Oberthuer D, Kikhney A, Guenther T, Fahmy K, Svergun D, Betzel C, Raff J
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
495 |
nm3 |
|
|