|
|
|
|
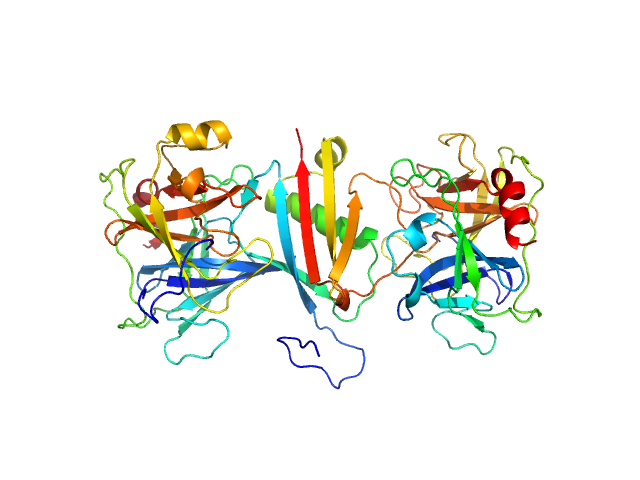
|
| Sample: |
Cathepsin G monomer, 25 kDa Homo sapiens protein
MAP domain-containing protein monomer, 13 kDa Staphylococcus aureus (strain … protein
Neutrophil elastase monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
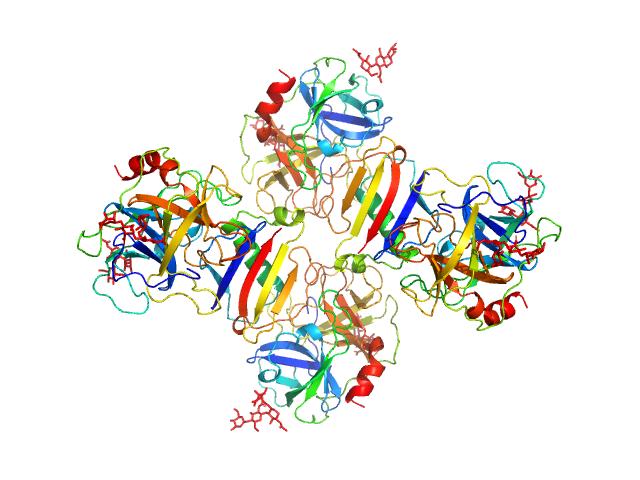
|
| Sample: |
Neutrophil elastase tetramer, 93 kDa Homo sapiens protein
Protein map dimer, 24 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 26
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
|
|
|
|
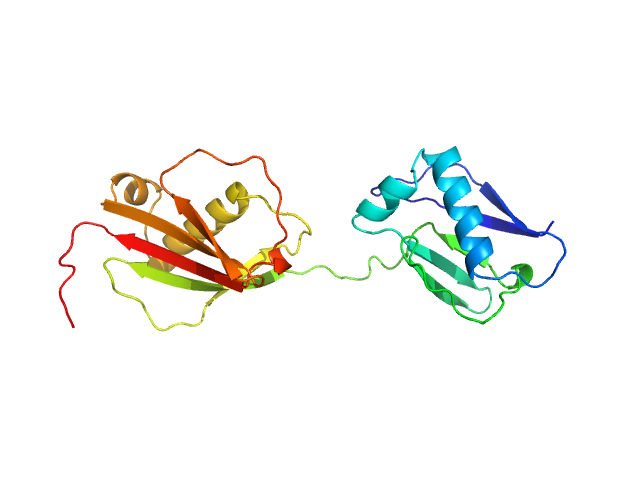
|
| Sample: |
Protein map monomer, 24 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Nov 17
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
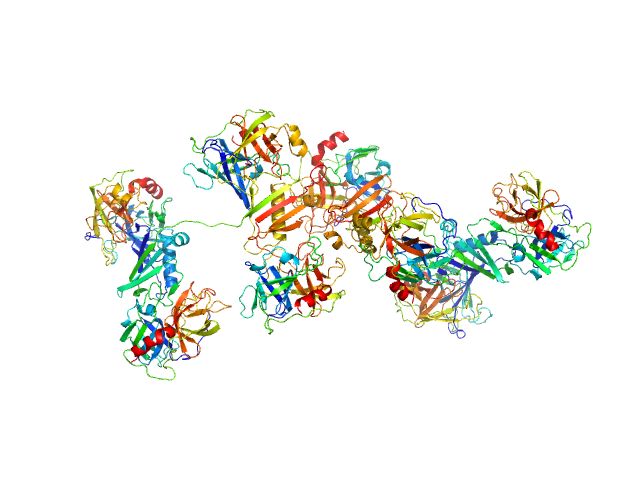
|
| Sample: |
Cathepsin G tetramer, 101 kDa Homo sapiens protein
Neutrophil elastase tetramer, 93 kDa Homo sapiens protein
Eap34 dimer, 48 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
500 |
nm3 |
|
|
|
|
|
|
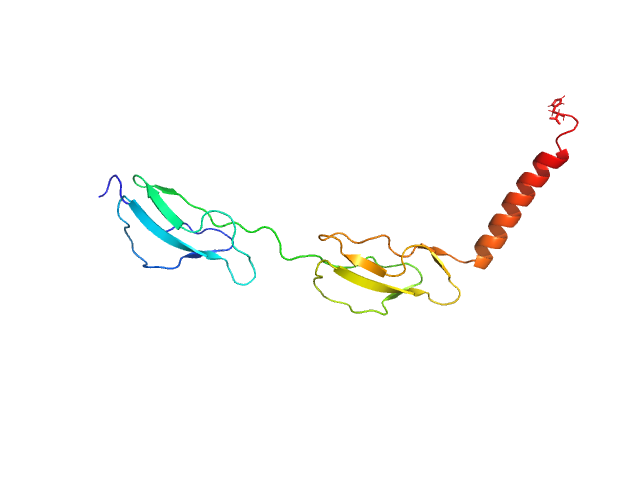
|
| Sample: |
Complement factor H monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 137 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 20
|
The SCR-17 and SCR-18 glycans in human complement Factor H enhance its regulatory function.
J Biol Chem :107624 (2024)
Gao X, Iqbal H, Yu DQ, Gor J, Coker AR, Perkins SJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
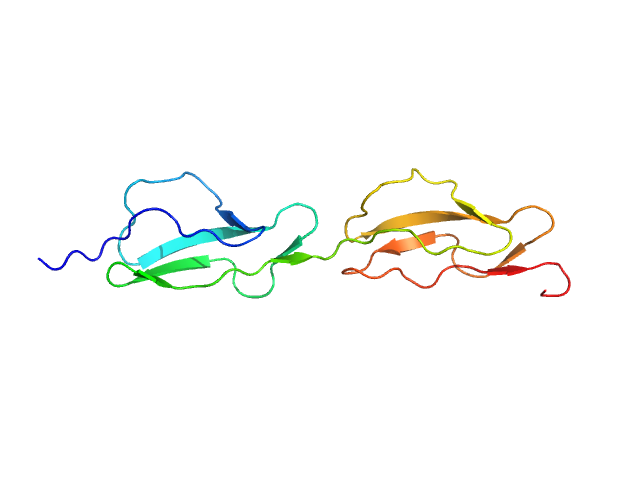
|
| Sample: |
Complement factor H monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 137 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 20
|
The SCR-17 and SCR-18 glycans in human complement Factor H enhance its regulatory function.
J Biol Chem :107624 (2024)
Gao X, Iqbal H, Yu DQ, Gor J, Coker AR, Perkins SJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
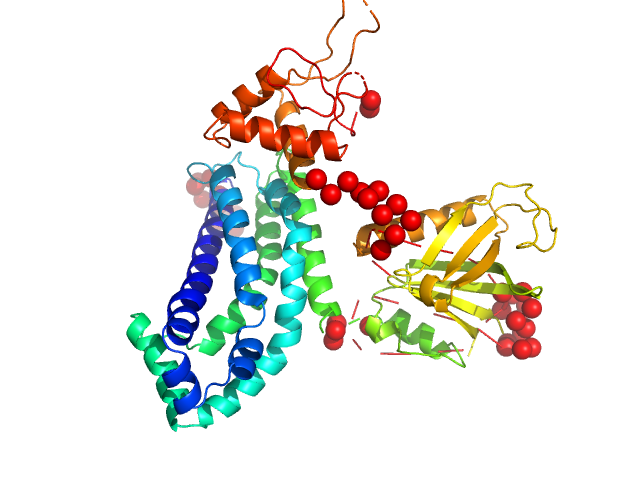
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
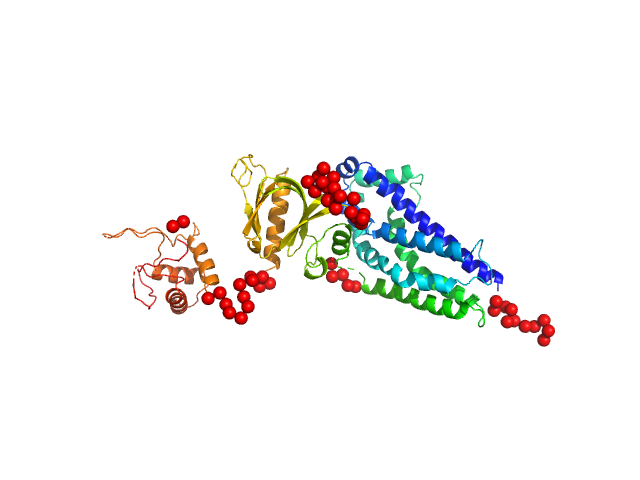
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (A170K) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 24
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
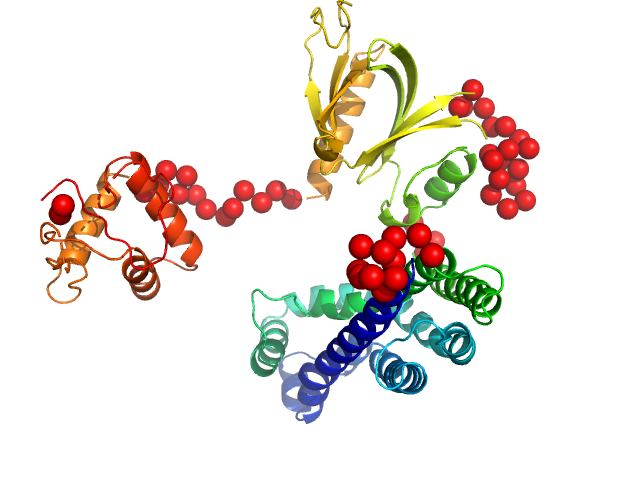
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (L177E) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
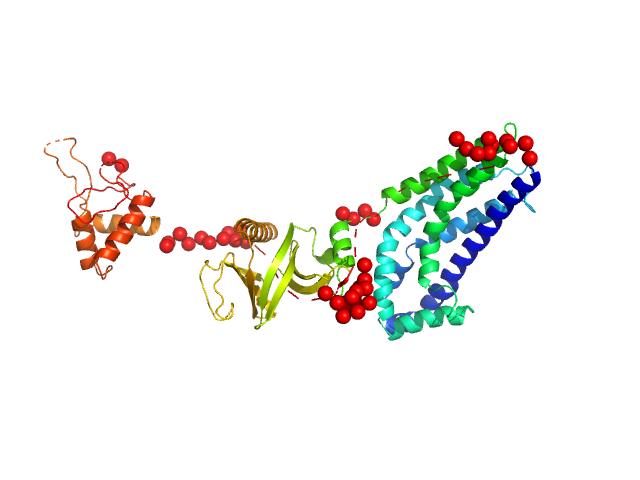
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (I409A) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
79 |
nm3 |
|
|