|
|
|
|

|
| Sample: |
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
13.4 |
nm |
| Dmax |
48.0 |
nm |
|
|
|
|
|
|
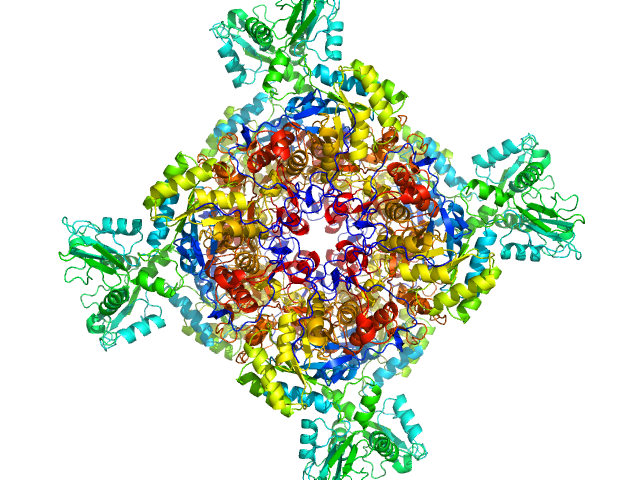
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Feb 21
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.3 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
952 |
nm3 |
|
|
|
|
|
|
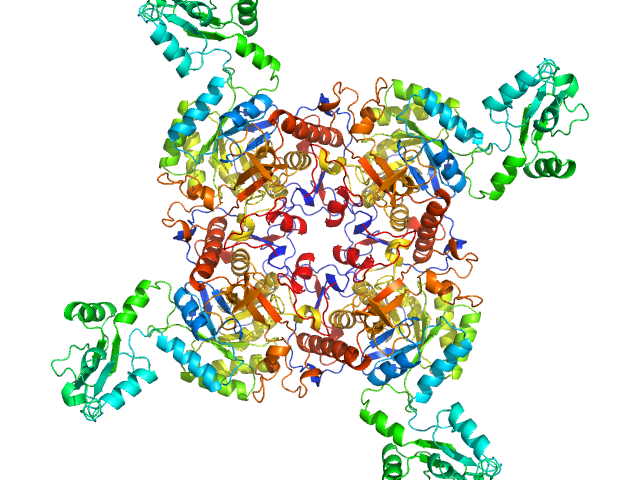
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase tetramer, 213 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
471 |
nm3 |
|
|
|
|
|
|
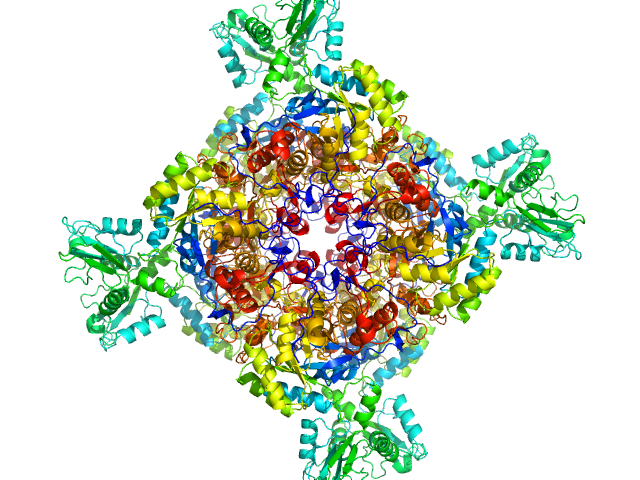
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Jan 29
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
821 |
nm3 |
|
|
|
|
|
|
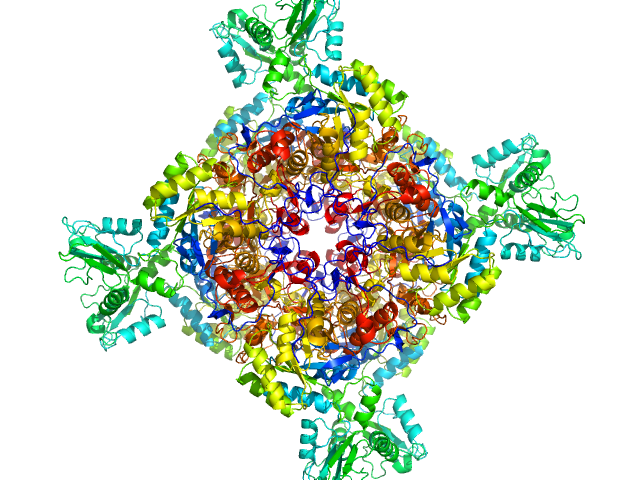
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.1 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
784 |
nm3 |
|
|
|
|
|
|
|
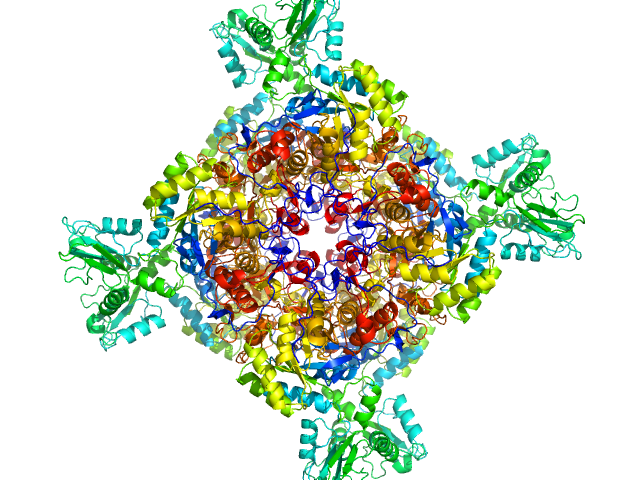
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
810 |
nm3 |
|
|
|
|
|
|
|
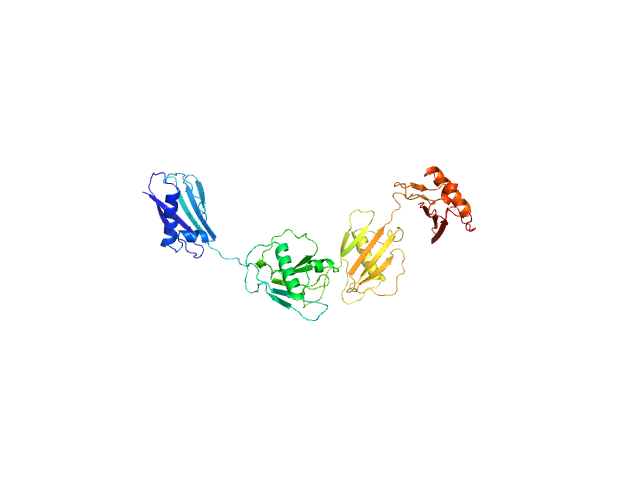
| Sample: |
Protein map monomer, 50 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
4.0 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
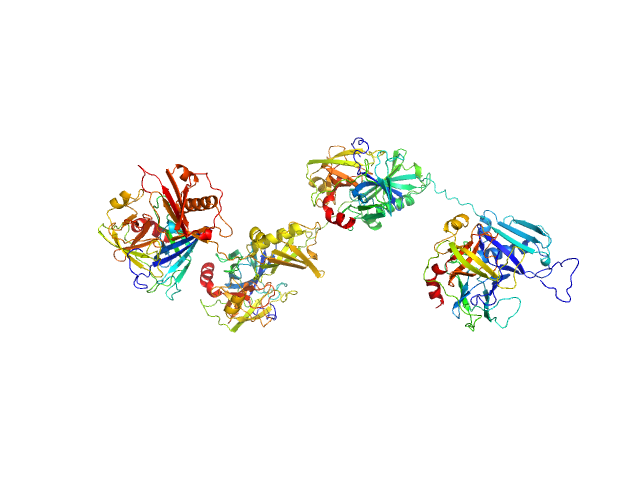
| Sample: |
Protein map monomer, 50 kDa Staphylococcus aureus (strain … protein
Cathepsin G tetramer, 101 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
|
|
|
|
|
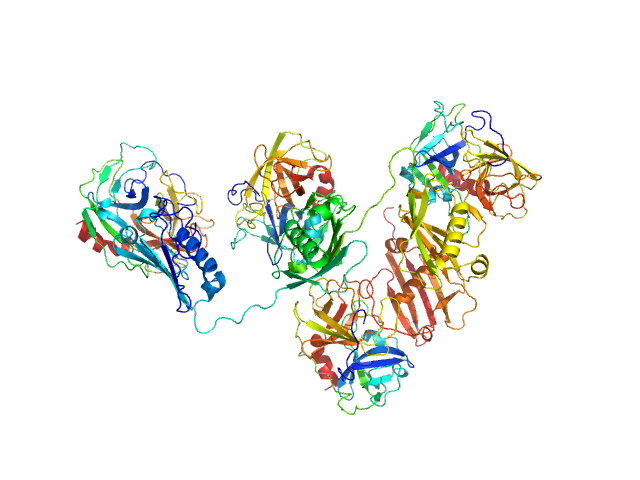
| Sample: |
Protein map monomer, 50 kDa Staphylococcus aureus (strain … protein
Neutrophil elastase tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Dec 6
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
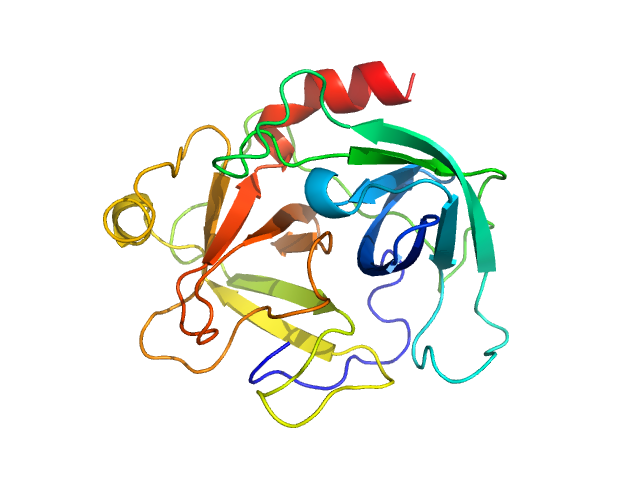
| Sample: |
Cathepsin G monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
27 |
nm3 |
|
|