|
|
|
|
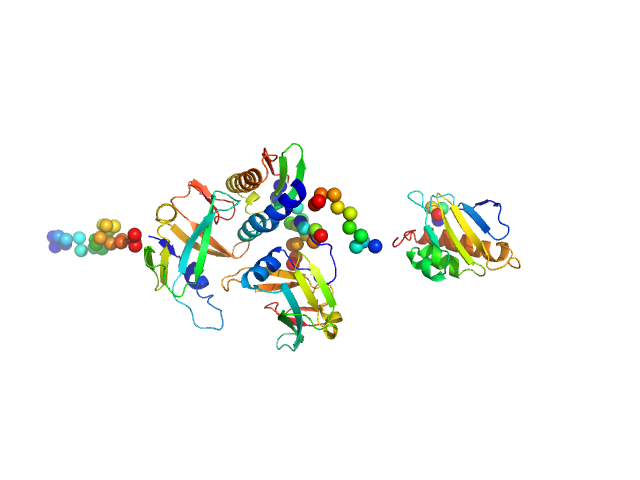
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G) monomer, 22 kDa Homo sapiens protein
Iron-sulfur cluster assembly 2 homolog, mitochondrial monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
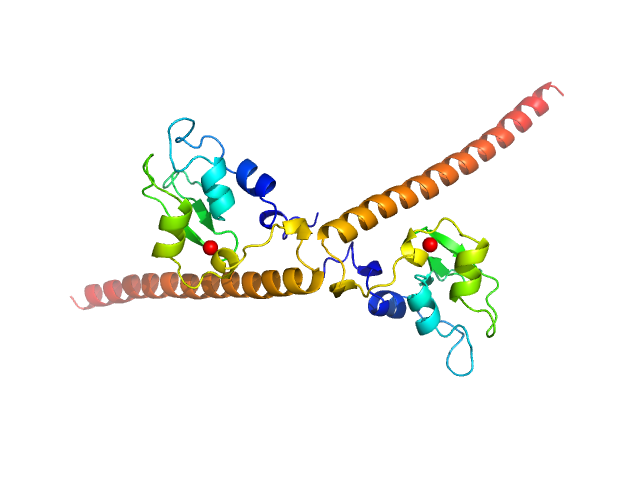
|
| Sample: |
Baculoviral IAP repeat-containing protein 5 dimer, 34 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 10
|
Survivin prevents the Polycomb Repressor Complex 2 from methylating Histone 3 lysine 27
iScience :106976 (2023)
Jensen M, Chandrasekaran V, García-Bonete M, Li S, Anindya A, Andersson K, Erlandsson M, Oparina N, Burmann B, Brath U, Panchenko A, Maria Bokarewa I, Katona G
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
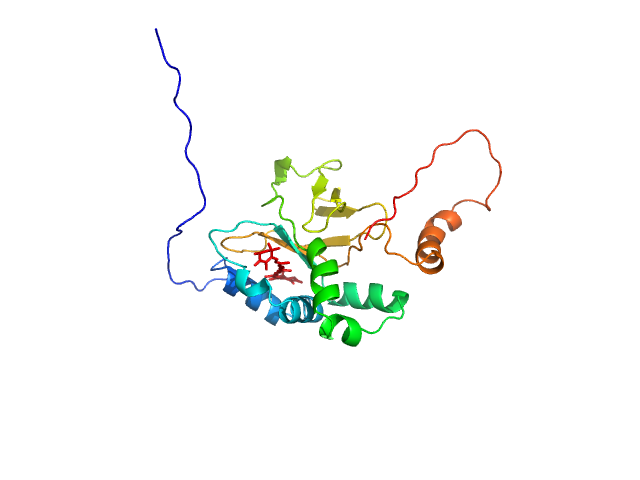
|
| Sample: |
Astaxanthin binding fasciclin family protein monomer, 22 kDa Coelastrella astaxanthina protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 24
|
Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol 6(1):471 (2023)
Kornilov FD, Slonimskiy YB, Lunegova DA, Egorkin NA, Savitskaya AG, Kleymenov SY, Maksimov EG, Goncharuk SA, Mineev KS, Sluchanko NN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
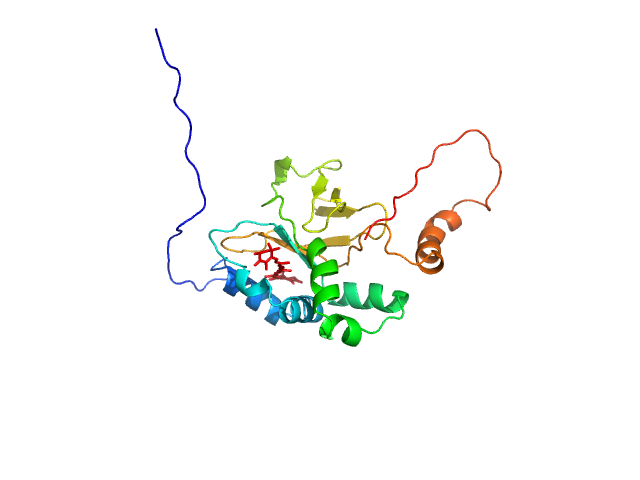
|
| Sample: |
Astaxanthin binding fasciclin family protein monomer, 22 kDa Coelastrella astaxanthina protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 24
|
Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol 6(1):471 (2023)
Kornilov FD, Slonimskiy YB, Lunegova DA, Egorkin NA, Savitskaya AG, Kleymenov SY, Maksimov EG, Goncharuk SA, Mineev KS, Sluchanko NN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
65 kDa invariant surface glycoprotein, putative monomer, 41 kDa Trypanosoma brucei gambiense … protein
Complement C3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Cryo-EM structures of Trypanosoma brucei gambiense ISG65 with human complement C3 and C3b and their roles in alternative pathway restriction.
Nat Commun 14(1):2403 (2023)
Sülzen H, Began J, Dhillon A, Kereïche S, Pompach P, Votrubova J, Zahedifard F, Šubrtova A, Šafner M, Hubalek M, Thompson M, Zoltner M, Zoll S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
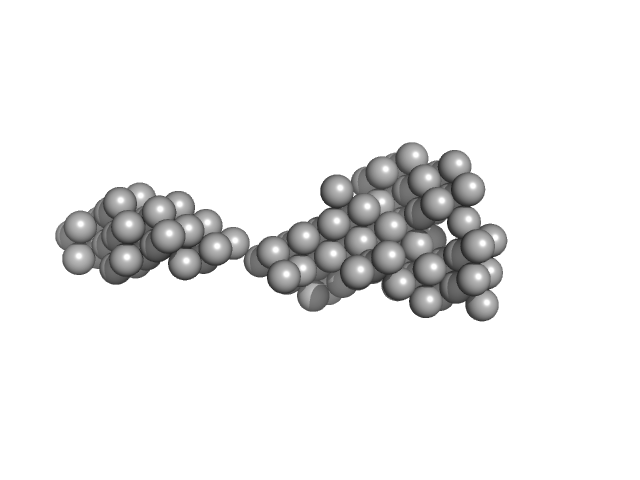
|
| Sample: |
Human Mucin 2 C-terminal dimer, 240 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 20
|
The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins
Nature Communications 14(1) (2023)
Gallego P, Garcia-Bonete M, Trillo-Muyo S, Recktenwald C, Johansson M, Hansson G
|
| RgGuinier |
8.4 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
684 |
nm3 |
|
|
|
|
|
|
|
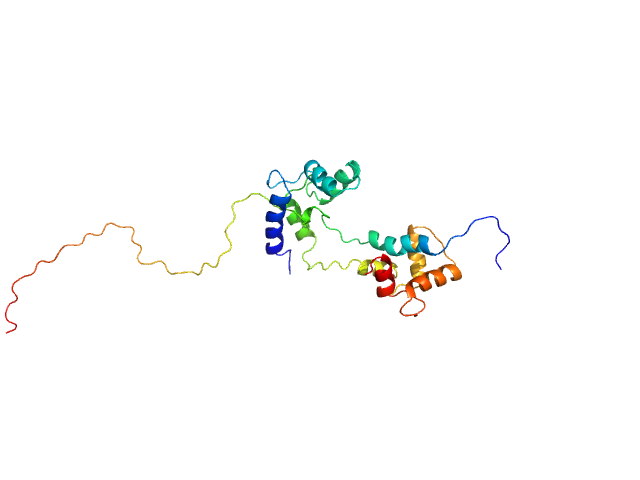
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Pre-mRNA-processing factor 40 homolog A monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Sep 23
|
Binding by calmodulin is coupled to transient unfolding of the third FF domain of Prp40A.
Protein Sci 32(4):e4606 (2023)
Díaz Casas A, Cordoba JJ, Ferrer BJ, Balakrishnan S, Wurm JE, Pastrana-Ríos B, Chazin WJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
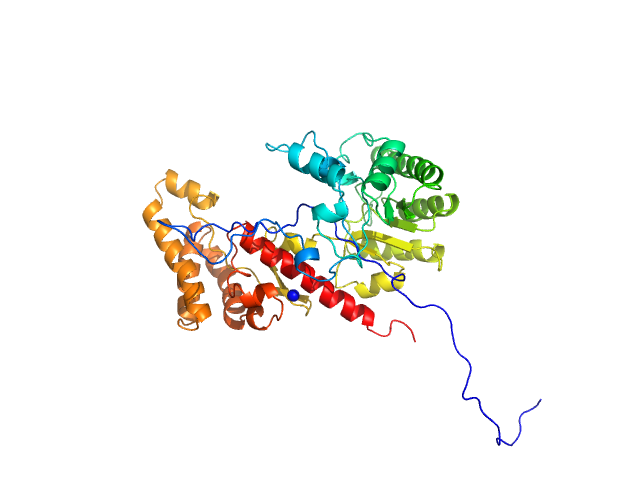
| Sample: |
DNA-guanine transglycosylase - D95A mutant monomer, 50 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
100 mM KCl, 50 mM Tris pH 7.0, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 22
|
7-Deazaguanines in DNA: functional and structural elucidation of a DNA modification system.
Nucleic Acids Res (2023)
Gedara SH, Wood E, Gustafson A, Liang C, Hung SH, Savage J, Phan P, Luthra A, de Crécy-Lagard V, Dedon P, Swairjo MA, Iwata-Reuyl D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
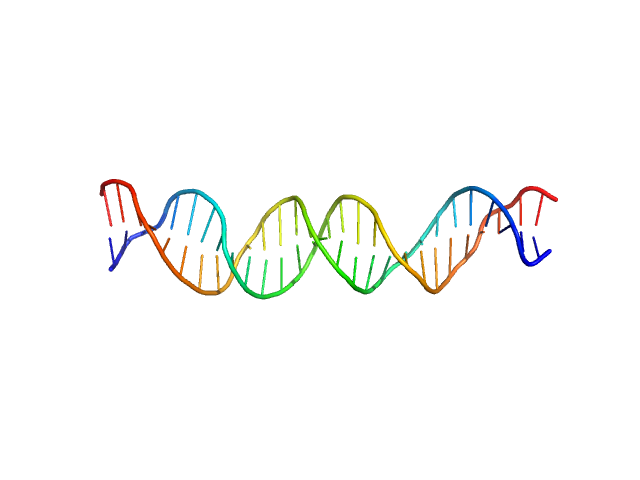
| Sample: |
Custom 28 base pair double stranded DNA monomer, 17 kDa Synthetic, purchased from … DNA
|
| Buffer: |
100 mM KCl, 50 mM Tris pH 7.0, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 22
|
7-Deazaguanines in DNA: functional and structural elucidation of a DNA modification system.
Nucleic Acids Res (2023)
Gedara SH, Wood E, Gustafson A, Liang C, Hung SH, Savage J, Phan P, Luthra A, de Crécy-Lagard V, Dedon P, Swairjo MA, Iwata-Reuyl D
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
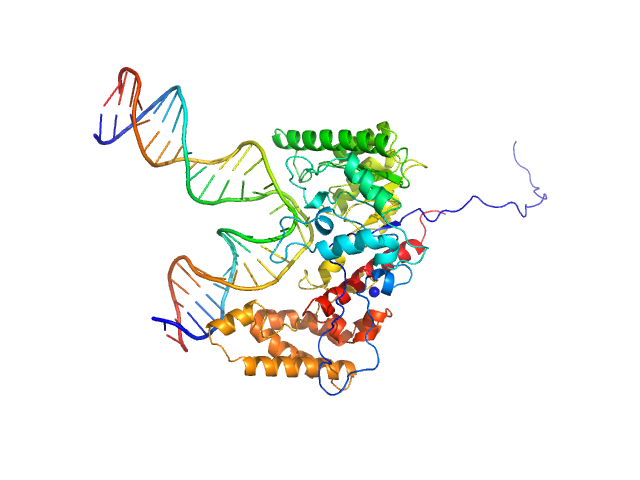
|
| Sample: |
Custom 28 base pair double stranded DNA monomer, 17 kDa Synthetic, purchased from … DNA
DNA-guanine transglycosylase - D95A mutant monomer, 50 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
100 mM KCl, 50 mM Tris pH 7.0, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 22
|
7-Deazaguanines in DNA: functional and structural elucidation of a DNA modification system.
Nucleic Acids Res (2023)
Gedara SH, Wood E, Gustafson A, Liang C, Hung SH, Savage J, Phan P, Luthra A, de Crécy-Lagard V, Dedon P, Swairjo MA, Iwata-Reuyl D
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
104 |
nm3 |
|
|