|
|
|
|
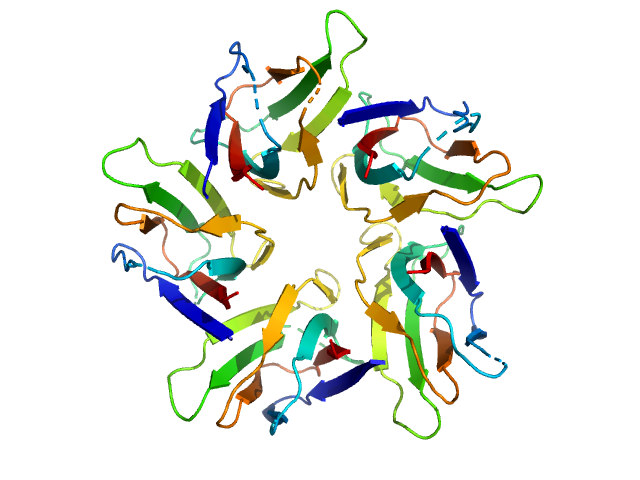
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 58 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 16
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
|
|
|
|
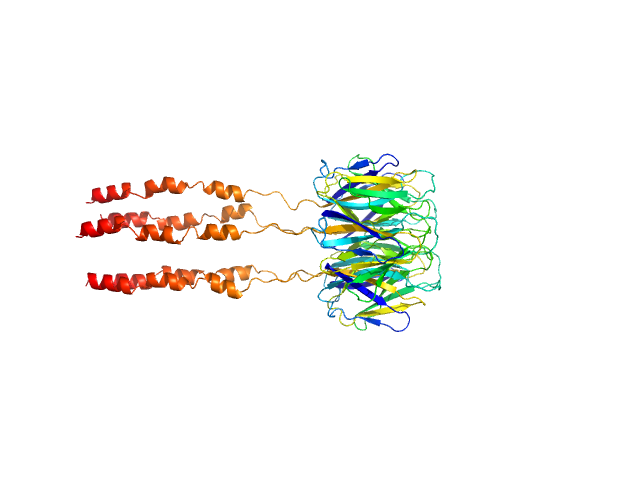
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 81 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
164 |
nm3 |
|
|
|
|
|
|
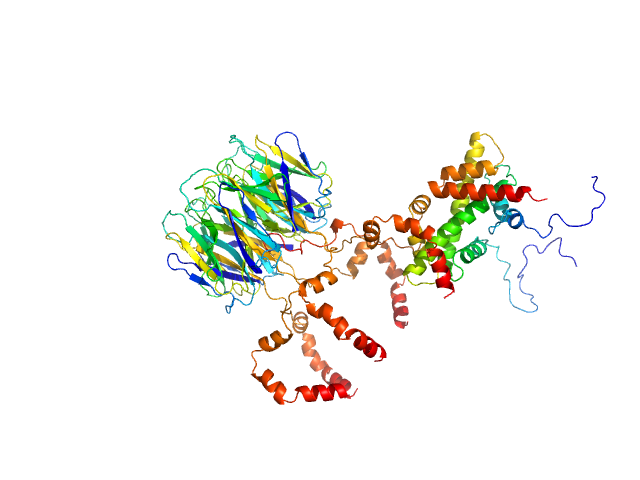
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 81 kDa Arabidopsis thaliana protein
Histone H2A type 1-B/E monomer, 14 kDa Homo sapiens protein
Histone H2B type 1-C/E/F/G/I monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|
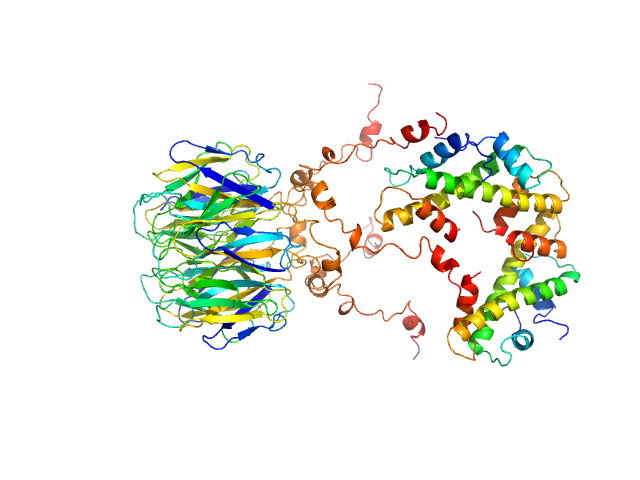
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 81 kDa Arabidopsis thaliana protein
Histone H3.1 dimer, 31 kDa Homo sapiens protein
Histone H4 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|
|
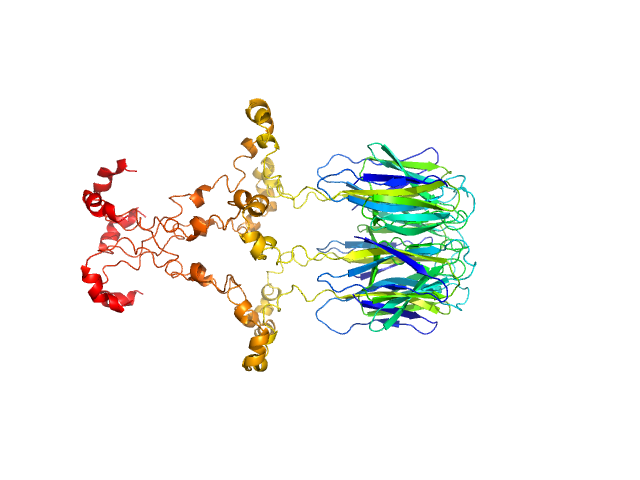
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 96 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
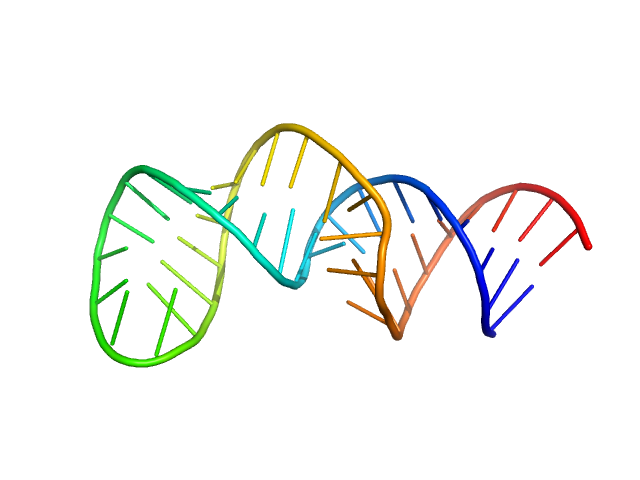
| Sample: |
The second exon splicing silencer 2p monomer, 14 kDa RNA
|
| Buffer: |
20 mM Bis-Tris, 20 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 1
|
Encoded Conformational Dynamics of the HIV Splice Site A3 Regulatory Locus: Implications for Differential Binding of hnRNP Splicing Auxiliary Factors.
J Mol Biol 434(18):167728 (2022)
Chiu LY, Emery A, Jain N, Sugarman A, Kendrick N, Luo L, Ford W, Swanstrom R, Tolbert BS
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
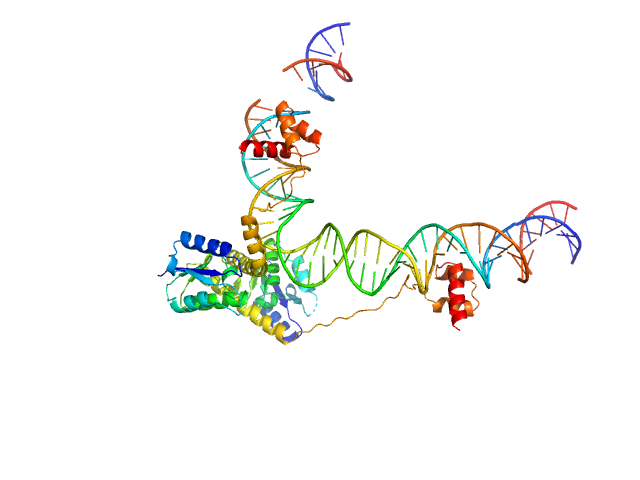
| Sample: |
Transposon Tn3 resolvase dimer, 43 kDa Escherichia coli protein
Tn3 res, resolvase binding site II monomer, 32 kDa DNA
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 10 mM MgCl2, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 6
|
Structural basis for topological regulation of Tn3 resolvase.
Nucleic Acids Res (2022)
Montaño SP, Rowland SJ, Fuller JR, Burke ME, MacDonald AI, Boocock MR, Stark WM, Rice PA
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
|
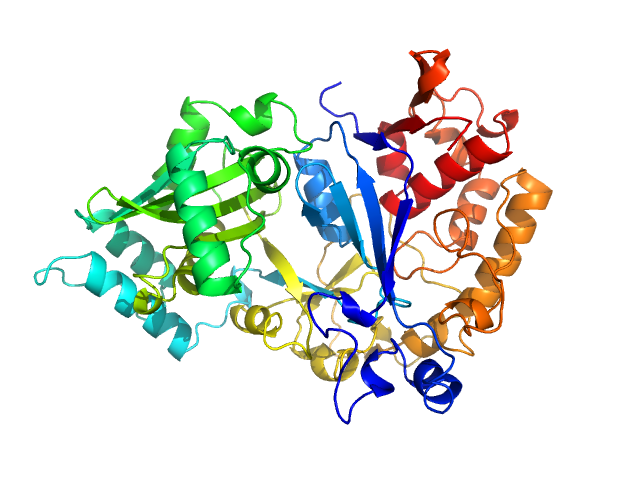
| Sample: |
Arginyl-tRNA--protein transferase 1 monomer, 60 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
2% glycerol, 100 mM KCl, 50 mM Tris-HCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 12
|
The Structure of Saccharomyces cerevisiae Arginyltransferase 1 (ATE1).
J Mol Biol 434(21):167816 (2022)
Van V, Ejimogu NE, Bui TS, Smith AT
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Testis-expressed protein 12 dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 10
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Testis-expressed protein 12 tetramer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 10
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|