|
|
|
|
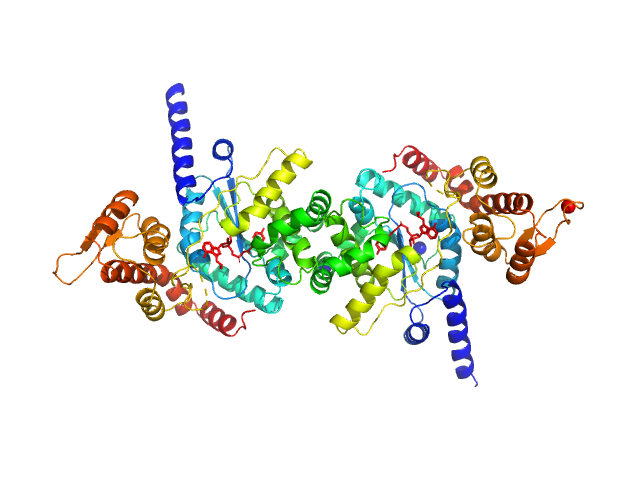
|
| Sample: |
Tyrosine--tRNA ligase (S234C) dimer, 87 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
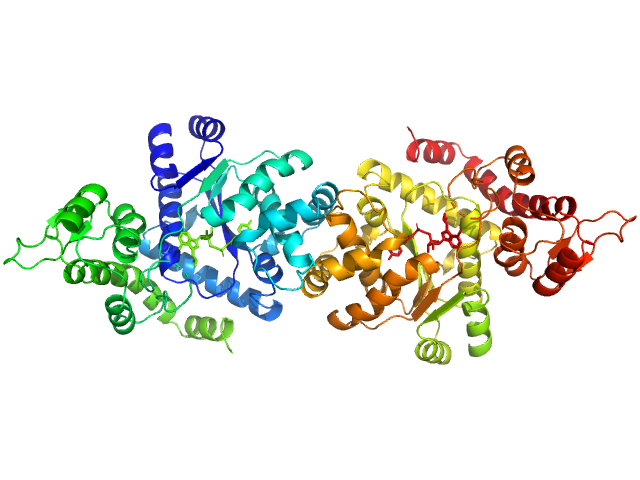
|
| Sample: |
Tyrosine--tRNA ligase, cytoplasmic dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Tegument protein UL21 (C-terminal domain) monomer, 29 kDa Human herpesvirus 1 … protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
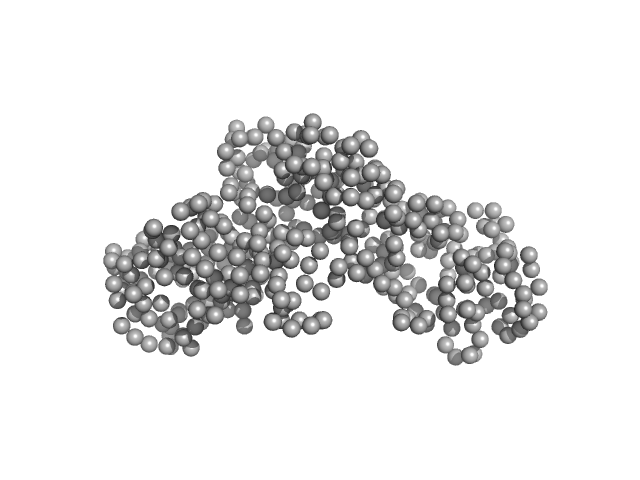
|
| Sample: |
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
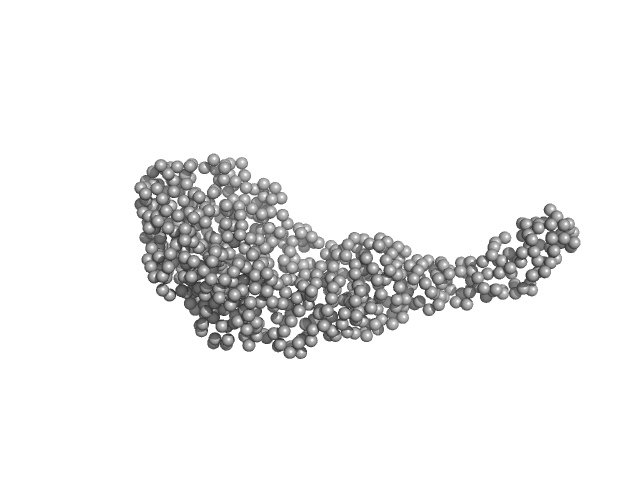
|
| Sample: |
Tegument protein UL21 (C-terminal domain) monomer, 29 kDa Human herpesvirus 1 … protein
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
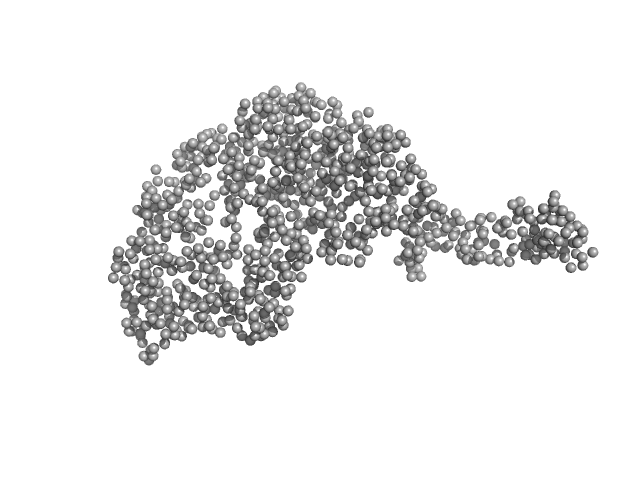
| Sample: |
Tegument protein UL21 monomer, 59 kDa Human alphaherpesvirus 1 … protein
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
|
|
|
|
|
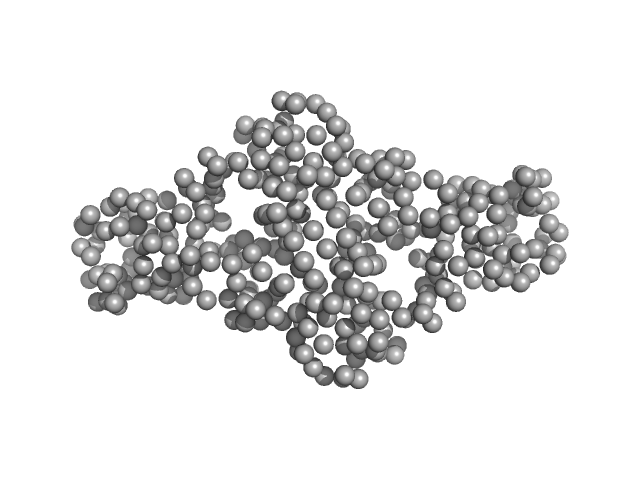
| Sample: |
Extracellular superoxide dismutase [Cu-Zn] dimer, 34 kDa Onchocerca volvulus protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 13
|
Crystal structure of an extracellular superoxide dismutase from Onchocerca volvulus
and implications for parasite-specific drug development
Acta Crystallographica Section F Structural Biology Communications 78(6) (2022)
Moustafa A, Perbandt M, Liebau E, Betzel C, Falke S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
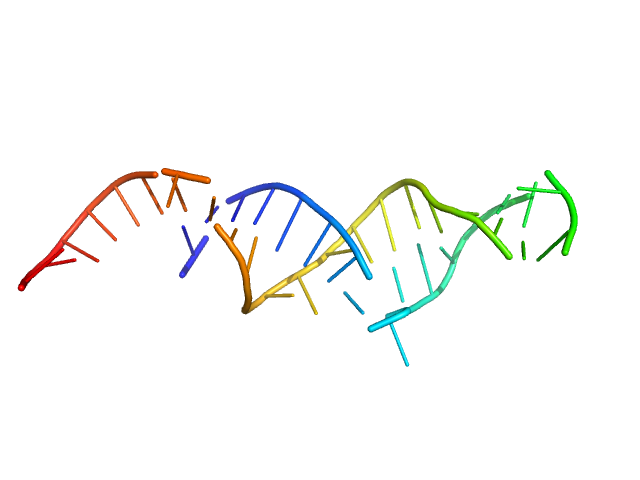
| Sample: |
Spiegelmer NOX-E36 monomer, 13 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
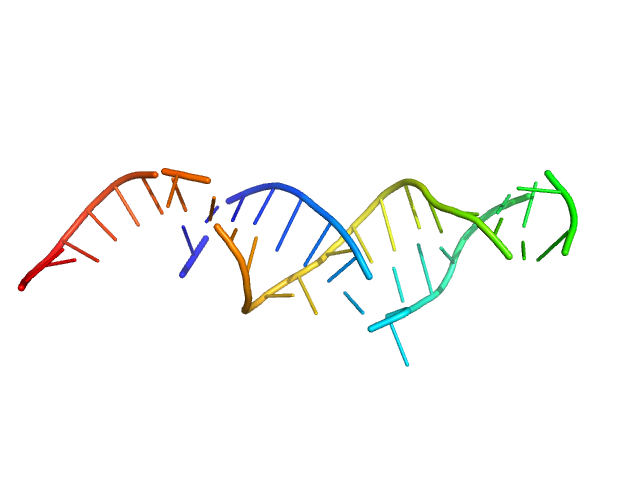
| Sample: |
Spiegelmer NOX-E36 monomer, 13 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
|
|
|
|
|
|
|
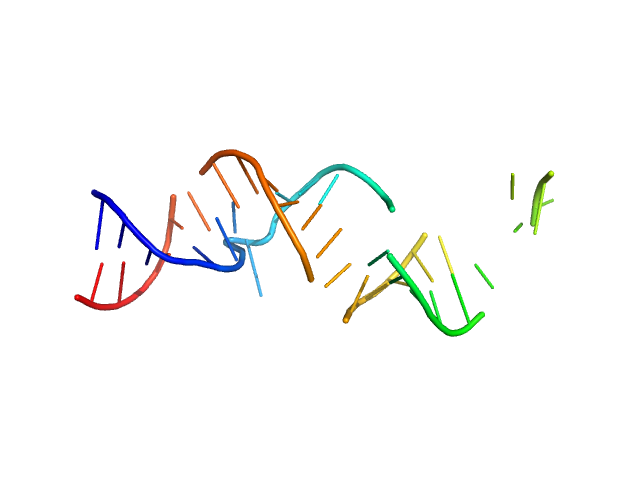
| Sample: |
Spiegelmer NOX-A12 monomer, 15 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
21 |
nm3 |
|
|