|
|
|
|
|
| Sample: |
Piwi domain-containing protein monomer, 91 kDa Pyrococcus furiosus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Piwi domain-containing protein monomer, 91 kDa Pyrococcus furiosus (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 250 mM NaCl, 2 mM DTT, 6 M urea, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Nov 17
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
148 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Clostridium butyricum argonaute protein monomer, 86 kDa protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Clostridium butyricum argonaute protein monomer, 86 kDa protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
|
|
|
|
|
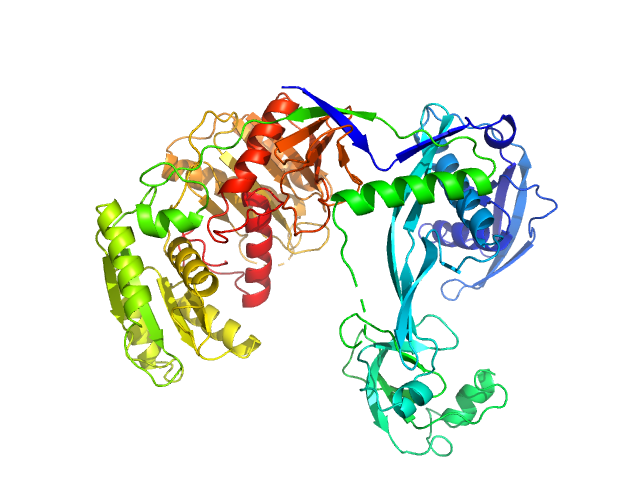
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|
|
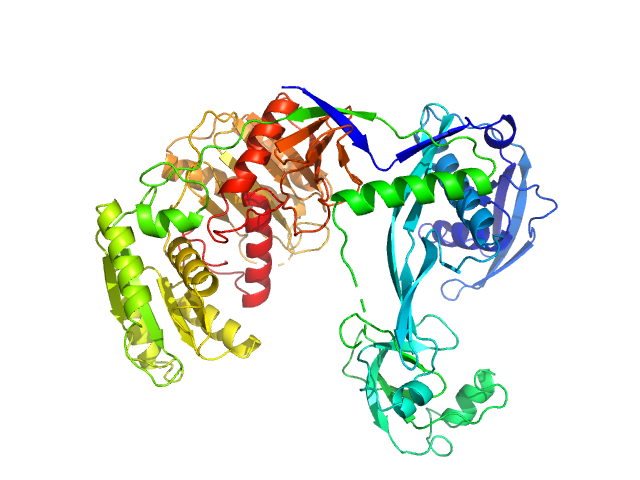
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
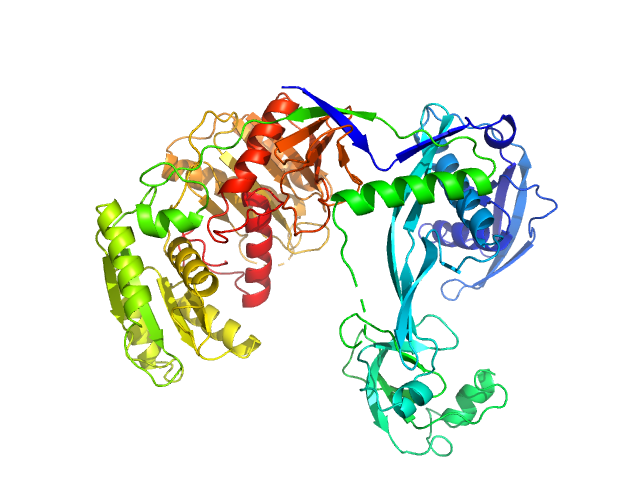
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
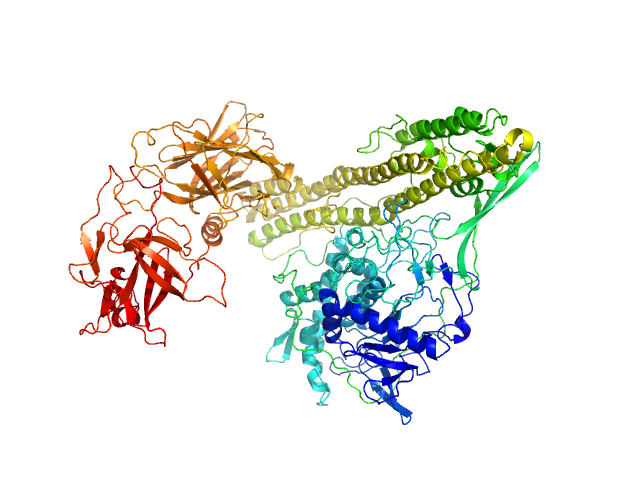
| Sample: |
Botulinum Toxin Serotype E, endonegative monomer, 144 kDa Clostridium botulinum protein
|
| Buffer: |
10 mM sodium acetate (20 mM ionic strength), pH: 4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 6
|
Elucidation of critical pH-dependent structural changes in Botulinum Neurotoxin E.
J Struct Biol :107876 (2022)
Lalaurie CJ, Splevins A, Barata TS, Bunting KA, Higazi DR, Zloh M, Spiteri VA, Perkins SJ, Dalby PA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
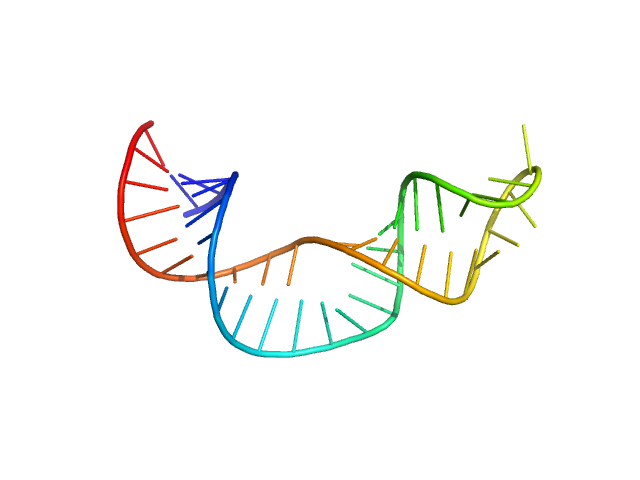
|
| Sample: |
Non-pre-microRNA stem loop 2 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Solution structure of NPSL2, a regulatory element in the oncomiR-1 RNA.
J Mol Biol :167688 (2022)
Liu Y, Munsayac A, Hall I, Keane SC
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
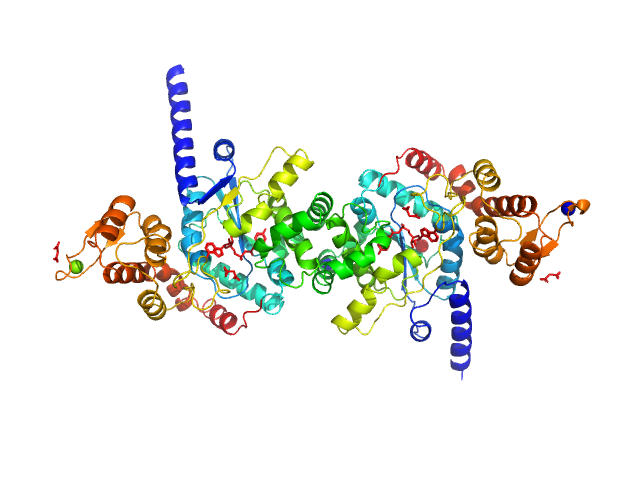
|
| Sample: |
Tyrosine--tRNA ligase dimer, 87 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
126 |
nm3 |
|
|