|
|
|
|
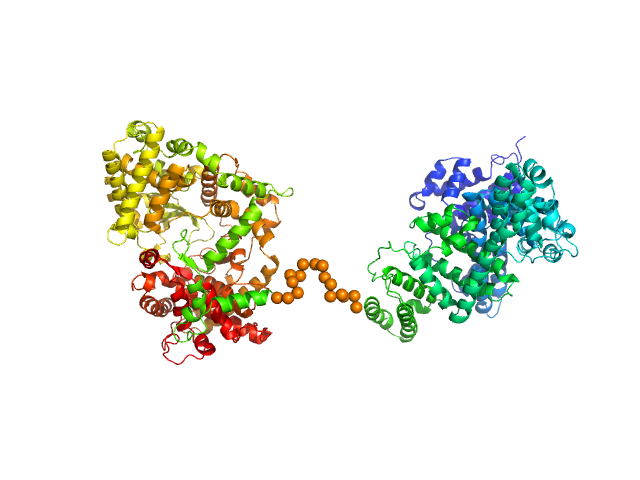
|
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM histidine, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 7
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
|
|
|
|
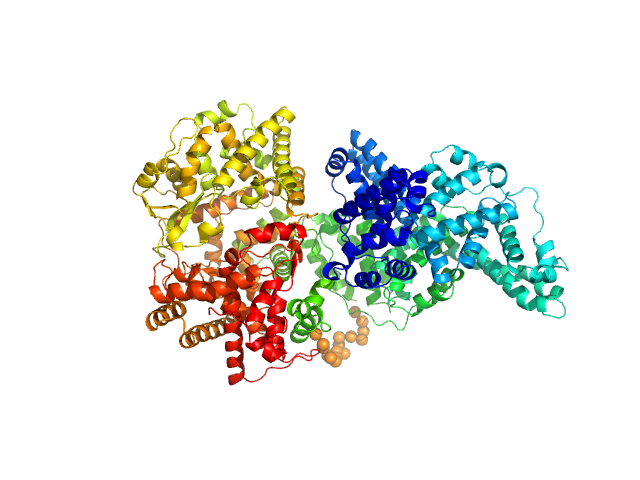
|
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM histidine, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 26
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
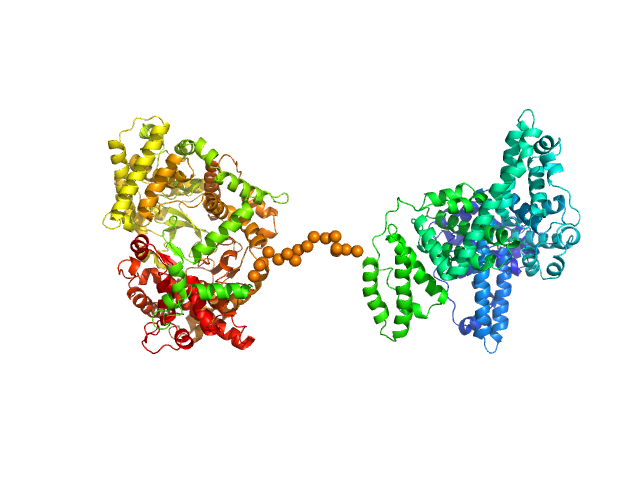
|
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM TRIS, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 15
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
239 |
nm3 |
|
|
|
|
|
|
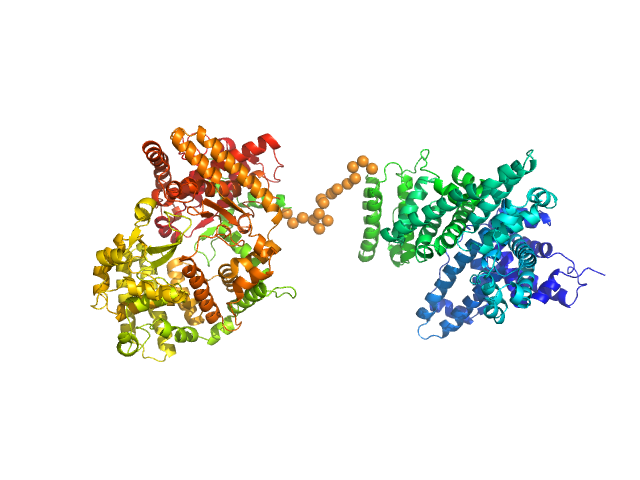
|
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 7
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
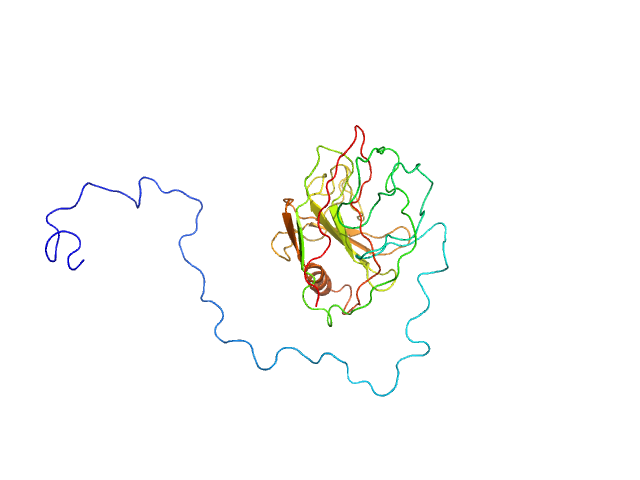
|
| Sample: |
Type III effector NopAA monomer, 31 kDa Sinorhizobium fredii USDA257 protein
|
| Buffer: |
PBS, 150 mM NaCl, 10% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 17
|
Structural and enzymatic characterisation of the Type III effector NopAA (=GunA) from Sinorhizobium fredii USDA257 reveals a Xyloglucan hydrolase activity.
Sci Rep 10(1):9932 (2020)
Dorival J, Philys S, Giuntini E, Brailly R, de Ruyck J, Czjzek M, Biondi E, Bompard C
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
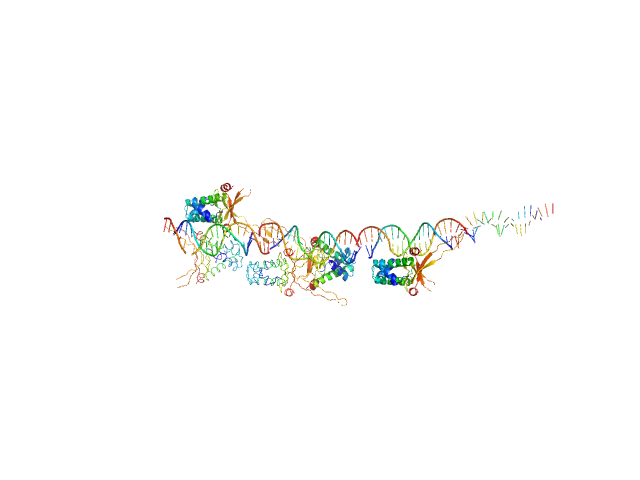
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 16-mer, 153 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 27
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
8.9 |
nm |
| Dmax |
28.5 |
nm |
| VolumePorod |
410 |
nm3 |
|
|
|
|
|
|
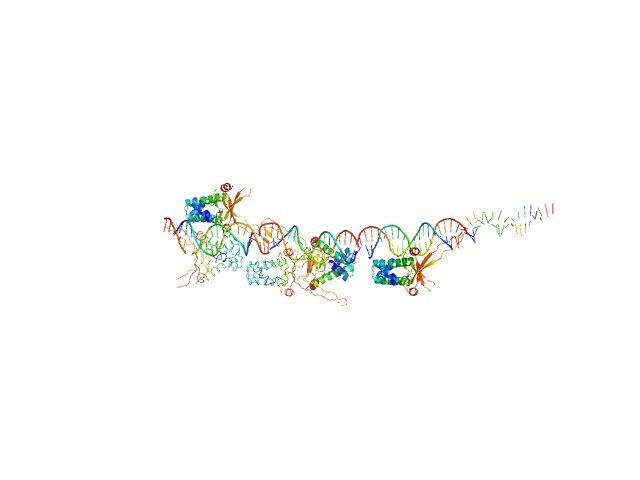
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 16-mer, 153 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
6.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
336 |
nm3 |
|
|
|
|
|
|
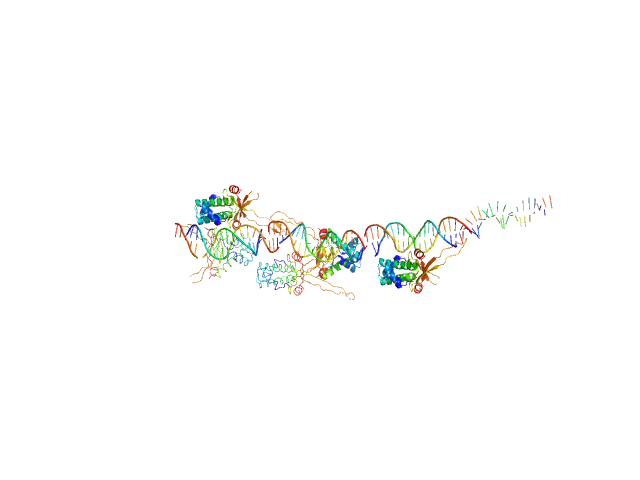
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 14-mer, 133 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
5.8 |
nm |
| Dmax |
24.2 |
nm |
| VolumePorod |
308 |
nm3 |
|
|
|
|
|
|
|
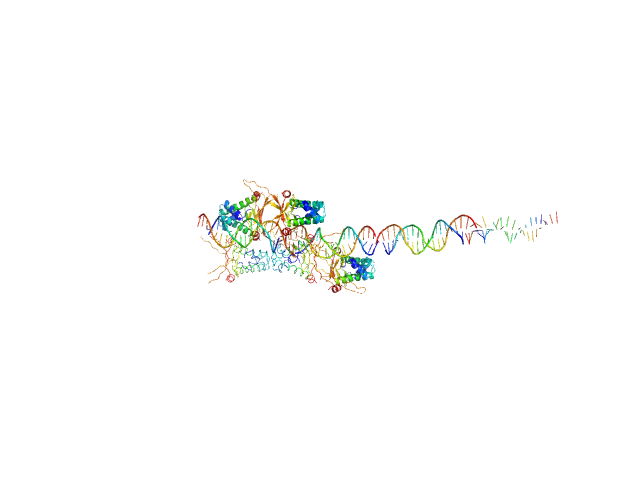
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha decamer, 95 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
6.5 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
242 |
nm3 |
|
|
|
|
|
|
|
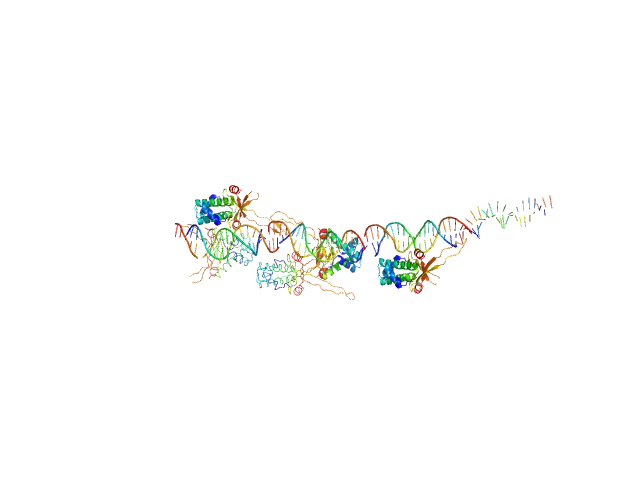
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 14-mer, 133 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.4 |
nm |
| VolumePorod |
274 |
nm3 |
|
|