|
|
|
|
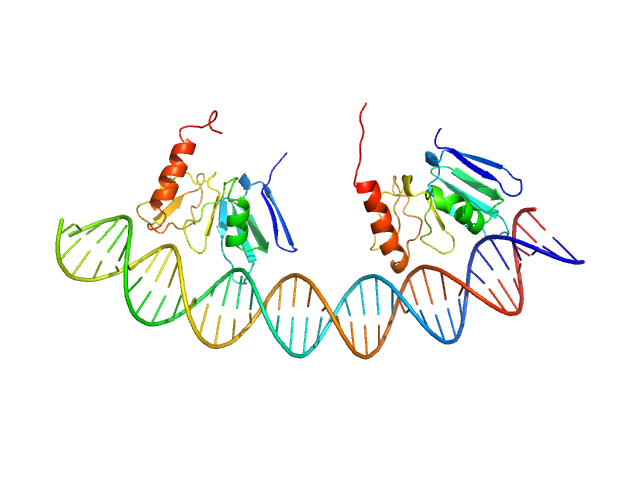
|
| Sample: |
Comcde, 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 29 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Jun 2
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Modification methylase SsoII monomer, 43 kDa Shigella sonnei protein
12-bp DNA monomer, 8 kDa DNA
|
| Buffer: |
50 mM Na-phosphate buffer, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Mar 13
|
Flexibility of the linker between the domains of DNA methyltransferase SsoII revealed by small-angle X-ray scattering: implications for transcription regulation in SsoII restriction-modification system.
PLoS One 9(4):e93453 (2014)
Konarev PV, Kachalova GS, Ryazanova AY, Kubareva EA, Karyagina AS, Bartunik HD, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.8 |
nm |
|
|
|
|
|
|
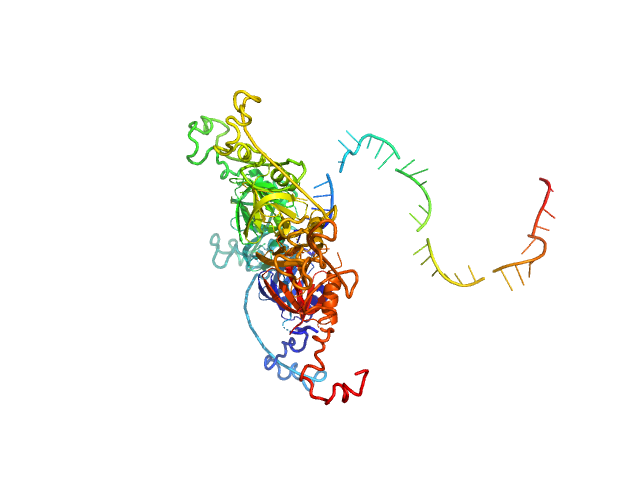
|
| Sample: |
RNA chaperone Hfq hexamer, 67 kDa Escherichia coli protein
RNA DsrA monomer, 12 kDa RNA
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 16
|
Structural flexibility of RNA as molecular basis for Hfq chaperone function.
Nucleic Acids Res 40(16):8072-84 (2012)
Ribeiro Ede A Jr, Beich-Frandsen M, Konarev PV, Shang W, Vecerek B, Kontaxis G, Hämmerle H, Peterlik H, Svergun DI, Bläsi U, Djinović-Carugo K
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
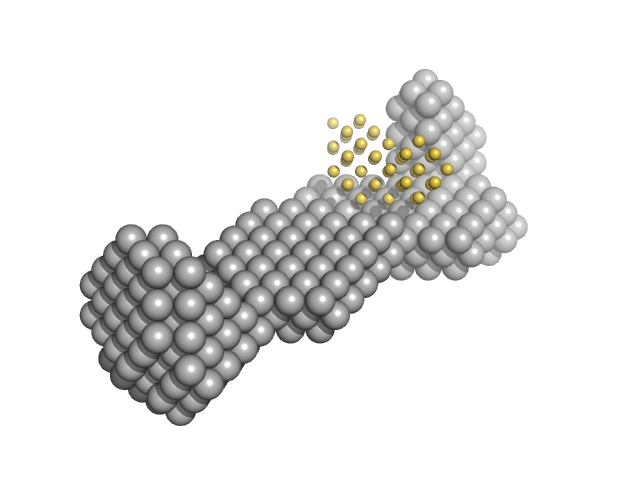
|
| Sample: |
Replication origin-binding protein dimer, 81 kDa Human herpesvirus 1 … protein
15-bp DNA monomer, 9 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 27
|
Structural and biophysical characterization of the proteins interacting with the herpes simplex virus 1 origin of replication.
J Biol Chem 284(24):16343-16353 (2009)
Manolaridis I, Mumtsidu E, Konarev P, Makhov AM, Fullerton SW, Sinz A, Kalkhof S, McGeehan JE, Cary PD, Griffith JD, Svergun D, Kneale GG, Tucker PA
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
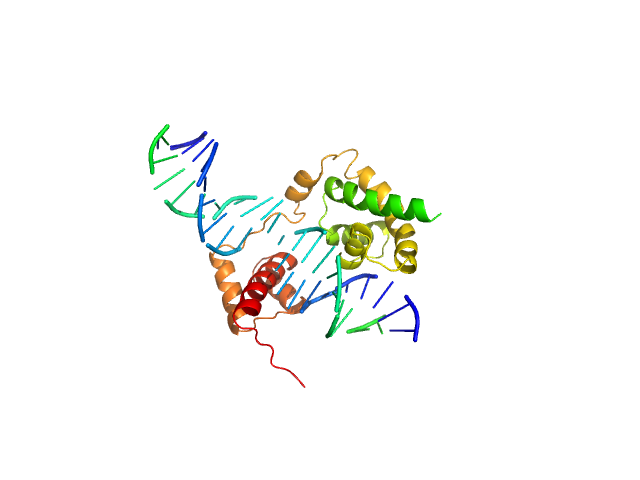
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Rat CRH DNA monomer, 15 kDa Rattus norvegicus DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2003 Oct 9
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
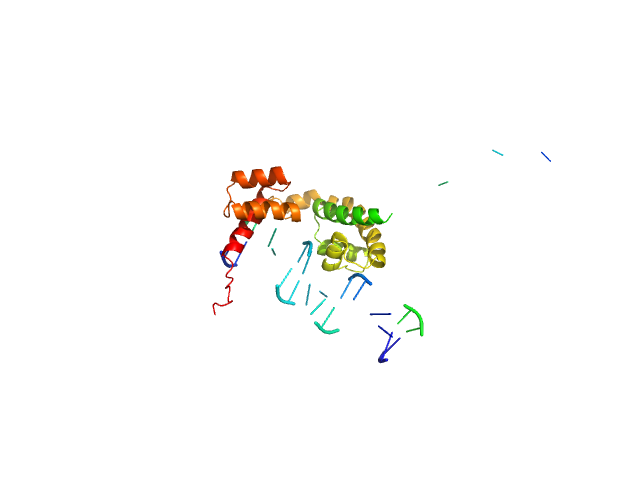
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Human DR-alpha DNA monomer, 15 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 May 26
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|