|
|
|
|

|
| Sample: |
Short-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 14 kDa Shewanella putrefaciens RNA
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Trimeric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) trimer, 112 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
Hexameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) hexamer, 223 kDa Shewanella putrefaciens protein
Wildtype-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 19 kDa RNA
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.4 |
nm |
|
|
|
|
|
|

|
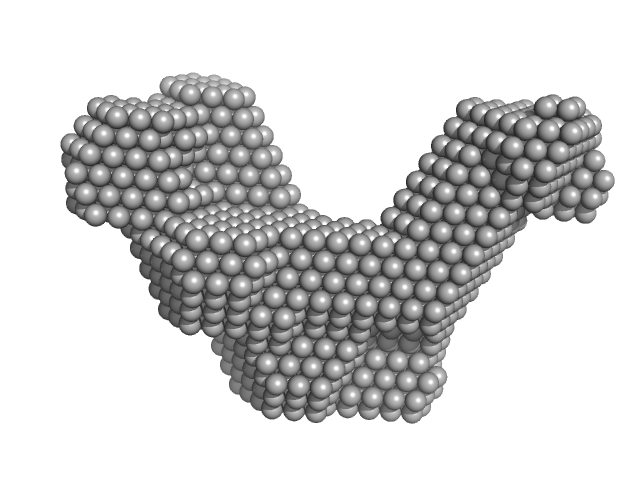
| Sample: |
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
Nonameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) nonamer, 335 kDa Shewanella putrefaciens protein
Long-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 25 kDa RNA
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jan 30
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
6.5 |
nm |
| Dmax |
21.6 |
nm |
|
|
|
|
|
|
|
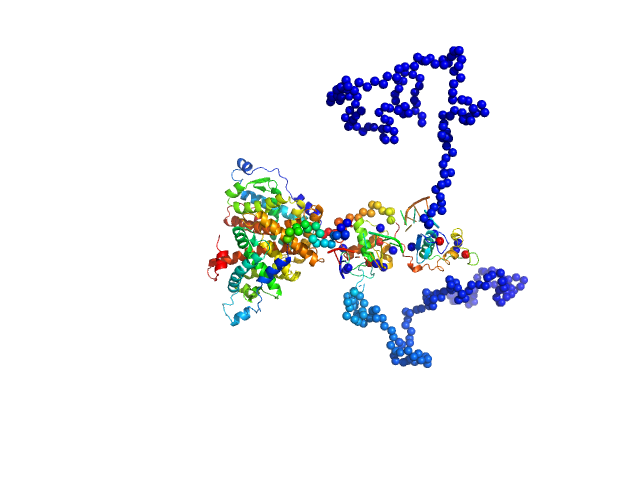
| Sample: |
Retinoic acid receptor RXR-alpha dimer, 102 kDa Homo sapiens protein
Ramp2 DNA monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 May 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|
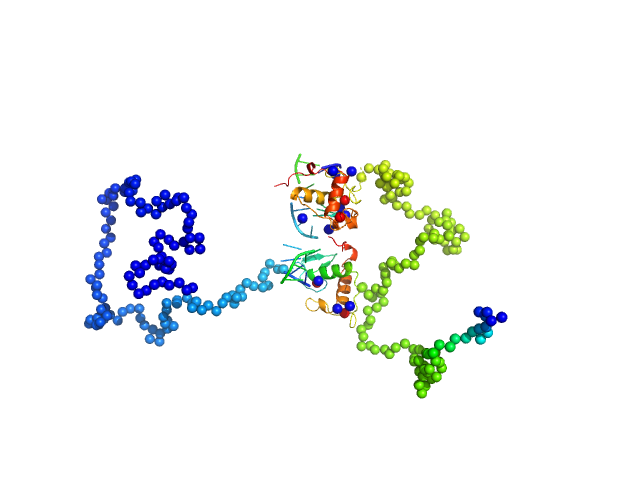
|
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Jul 17
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
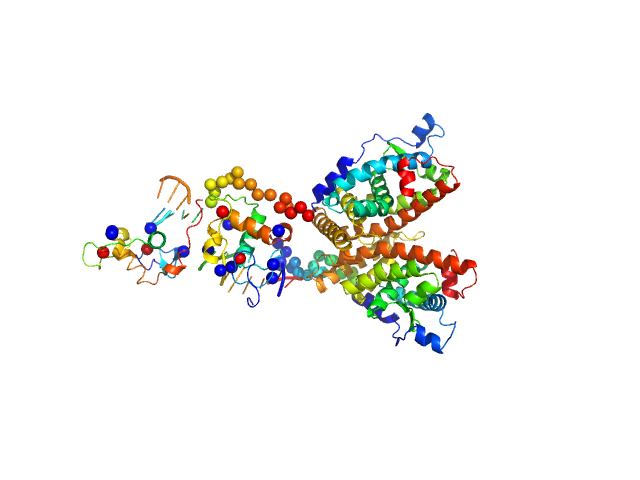
|
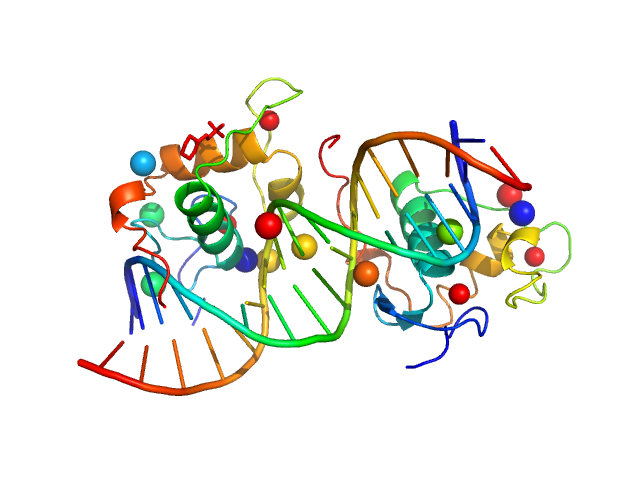
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 74 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
MHV-68 TR DNA monomer, 30 kDa unidentified herpesvirus DNA
Latency-associated nuclear antigen octamer, 269 kDa Murid herpesvirus 4 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
5.8 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
475 |
nm3 |
|
|
|
|
|
|

|
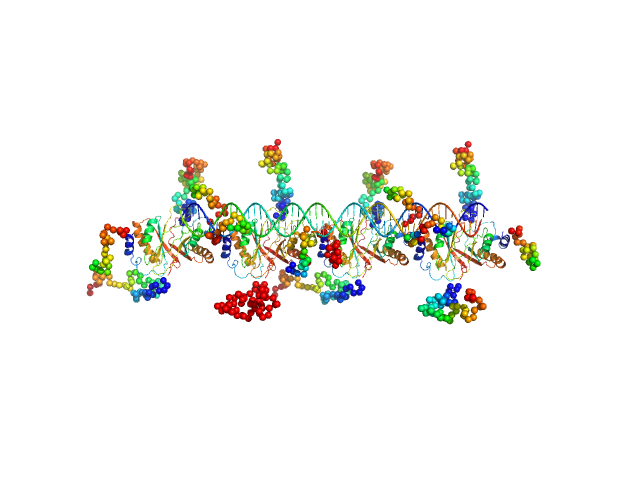
| Sample: |
KLBS1-2 DNA monomer, 24 kDa unidentified herpesvirus DNA
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 octamer octamer, 126 kDa Human herpesvirus 8 protein
KLBS1-2 DNA two monomers dimer, 48 kDa unidentified herpesvirus RNA
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|
|
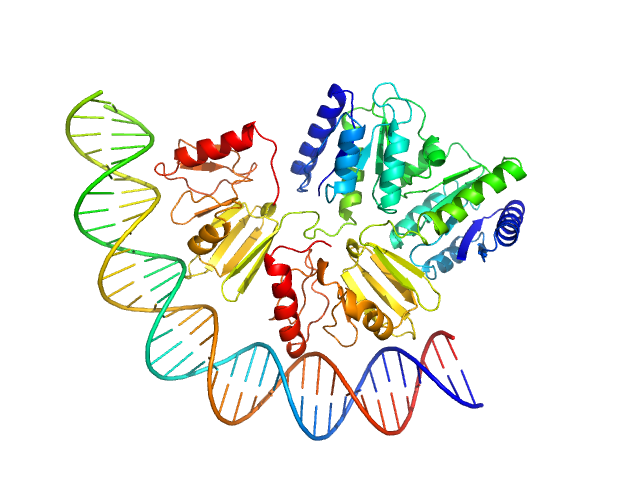
| Sample: |
Comcde, 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 61 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Feb 11
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
122 |
nm3 |
|
|