|
|
|
|
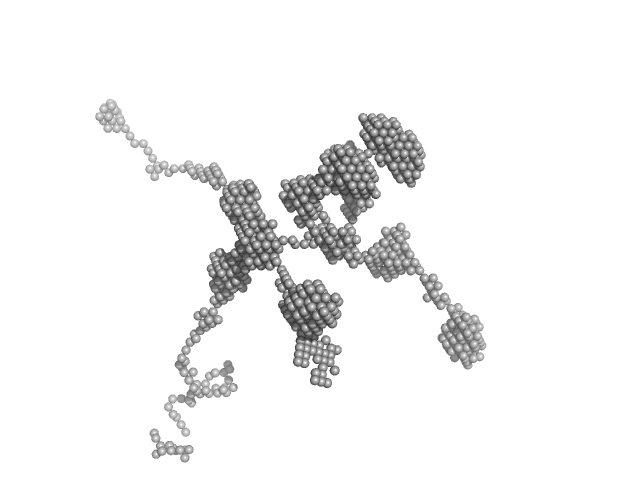
|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
90.5 |
nm |
| Dmax |
200.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
30.7 |
nm |
| Dmax |
107.4 |
nm |
| VolumePorod |
112000 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
Linear polyethylenimine monomer, 25 kDa none (polymer)
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
12.0 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
9100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LNA-DNA-LNA monomer, 4 kDa DNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MOE PS gapmers 3-8-3 monomer, 4 kDa DNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
OME ASO monomer, 4 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LNA-DNA-LNA monomer, 4 kDa DNA
PSCK9, 24mer ASO binding site monomer, 8 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MOE PS gapmers 3-8-3 monomer, 4 kDa DNA
PSCK9, 24mer ASO binding site monomer, 8 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
OME ASO monomer, 4 kDa RNA
PSCK9, 24mer ASO binding site monomer, 8 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
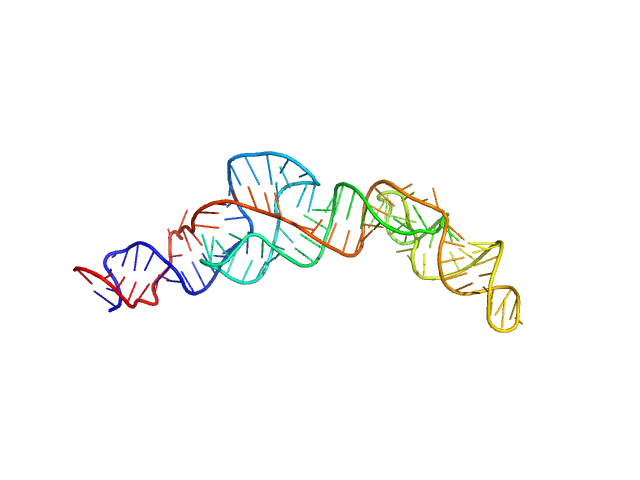
|
| Sample: |
RNA Device 43 in ligand-free state monomer, 40 kDa synthetic construct RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM KCl, and 1 mM MgCl2, pH: 6.8 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2023 Jul 25
|
Structural investigation of an RNA device that regulates PD-1 expression in mammalian cells.
Nucleic Acids Res 53(5) (2025)
Stagno JR, Deme JC, Dwivedi V, Lee YT, Lee HK, Yu P, Chen SY, Fan L, Degenhardt MFS, Chari R, Young HA, Lea SM, Wang YX
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|