|
|
|
|
|
| Sample: |
Lipid None, lipid
|
| Buffer: |
, pH: |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 19
|
Lipid_unpublished_76
Siawosch Schewa
|
|
|
|
|
|
|
|
| Sample: |
Lipid None, lipid
|
| Buffer: |
, pH: |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 19
|
Lipid_unpublished_77
Siawosch Schewa
|
|
|
|
|
|
|
|
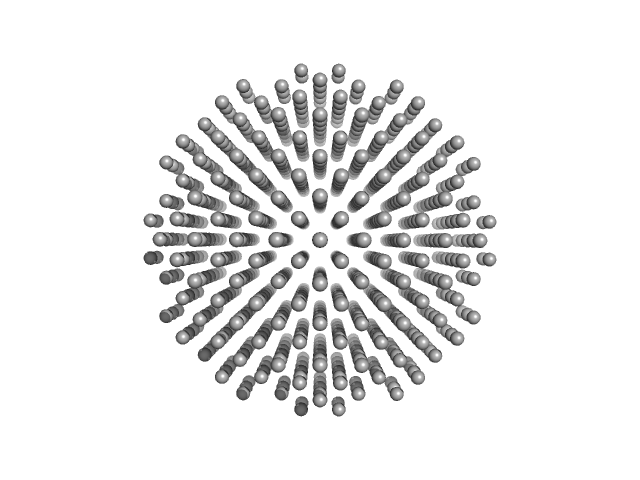
| Sample: |
Nominal 100 nm diameter polystyrene spheres monomer, 315312 kDa
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 16
|
Standard polystyrene nanospheres (concentration series data)
Dima Molodenskiy
|
| RgGuinier |
33.0 |
nm |
| Dmax |
100.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nominal 20 nm diameter polystyrene spheres 0, 8 kDa
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 16
|
Standard polystyrene nanospheres (concentration series data)
Dima Molodenskiy
|
| RgGuinier |
8.5 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
|
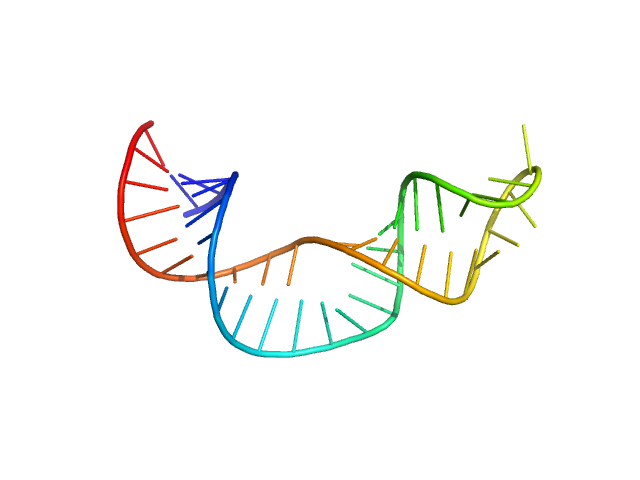
| Sample: |
Non-pre-microRNA stem loop 2 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Solution structure of NPSL2, a regulatory element in the oncomiR-1 RNA.
J Mol Biol :167688 (2022)
Liu Y, Munsayac A, Hall I, Keane SC
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
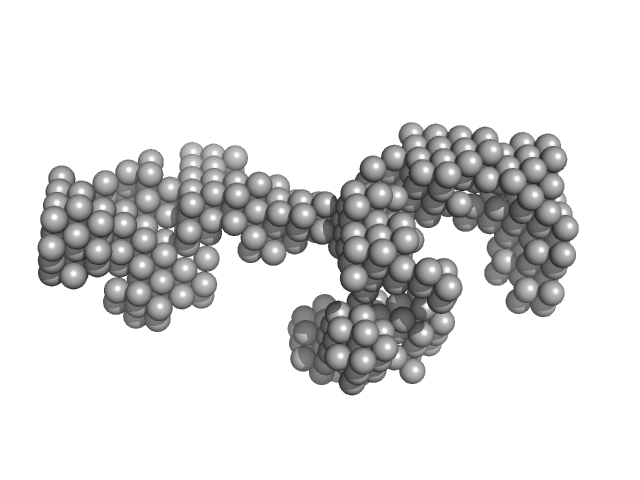
| Sample: |
LincRNA-p21 AluSx1 Sense RNA monomer, 100 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
6.1 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
|
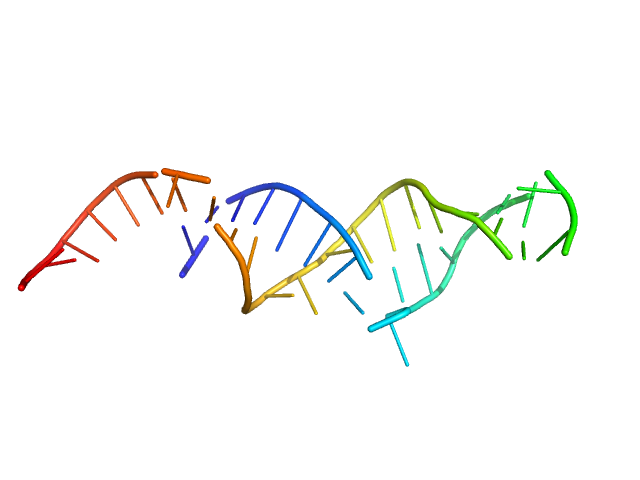
| Sample: |
LincRNA-p21 AluSx1 Antisense RNA monomer, 90 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
5.9 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
375 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Spiegelmer NOX-E36 monomer, 13 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
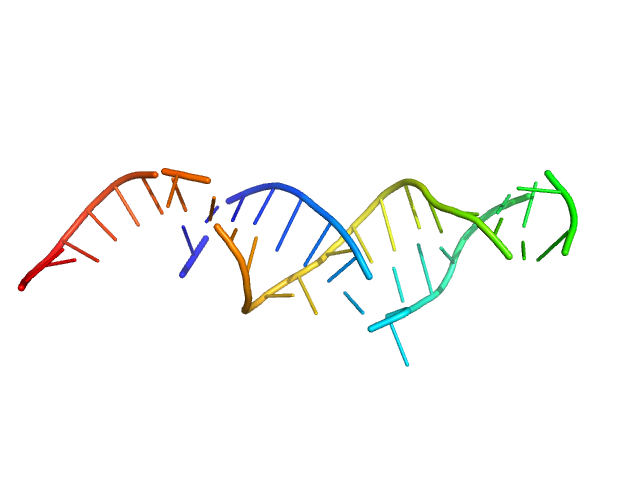
| Sample: |
Spiegelmer NOX-E36 monomer, 13 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
|
|
|
|
|
|
|
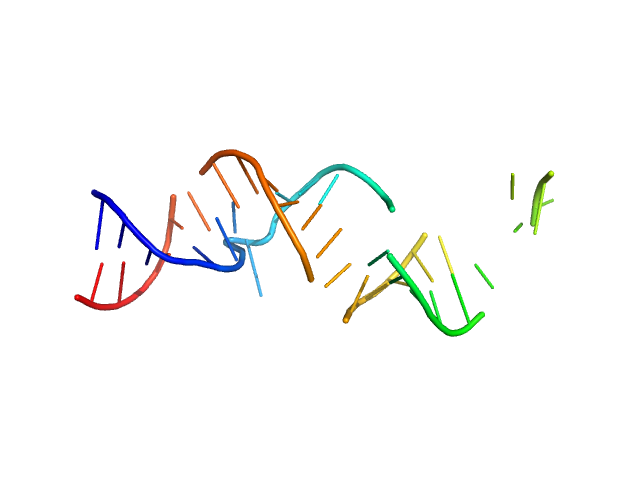
| Sample: |
Spiegelmer NOX-A12 monomer, 15 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
21 |
nm3 |
|
|