|
|
|
|
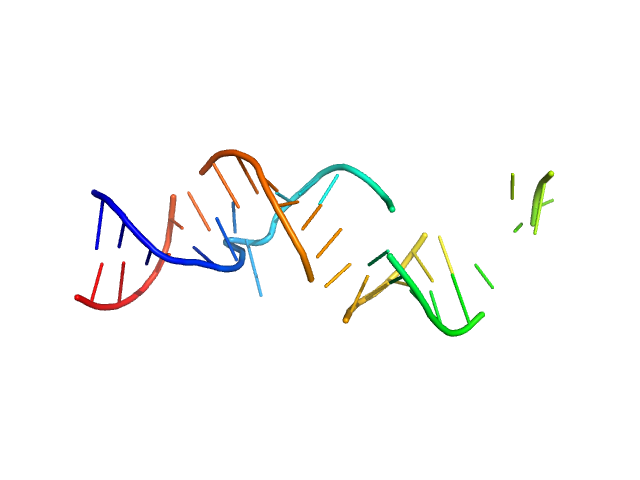
|
| Sample: |
Spiegelmer NOX-A12 monomer, 15 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 20
|
Spiegelmers® NOX-E36 and NOX-A12
Al Kikhney,
Bara Schmidt
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
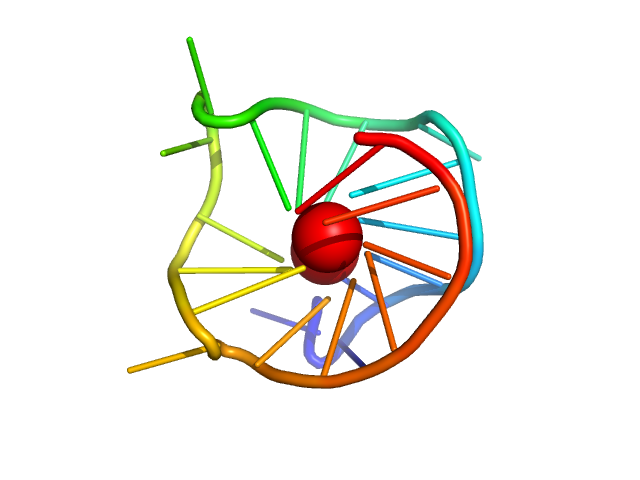
|
| Sample: |
MYC22-G14T/G23T monomer, 7 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 30 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Feb 27
|
Small-angle X-ray scattering data of a guanine-rich DNA derived from the promoter region of c-MYC gene in solution
Data in Brief :108285 (2022)
Miyauchi K, Imamura H, Yamaoki Y, Kato M
|
| RgGuinier |
1.3 |
nm |
| Dmax |
5.3 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
MYC22-G14T/G23T monomer, 7 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 100 mM 18-crown-6, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Feb 27
|
Small-angle X-ray scattering data of a guanine-rich DNA derived from the promoter region of c-MYC gene in solution
Data in Brief :108285 (2022)
Miyauchi K, Imamura H, Yamaoki Y, Kato M
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3'UTR full-length monomer, 50 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR ADE monomer, 16 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR CDE monomer, 9 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR ADE-CDE monomer, 24 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR Bulge monomer, 11 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR Bulge-ADE monomer, 27 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ox40 3 UTR Bulge-ADE-CDE monomer, 35 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 1
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|