|
|
|
|
|
| Sample: |
Β-chitin nanofibers from squid pens None, Squid
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5 |
| Experiment: |
SANS
data collected at BT5 USANS, NIST Center for Neutron Research on 2020 Dec 4
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
|
|
|
|
|
|
|
|
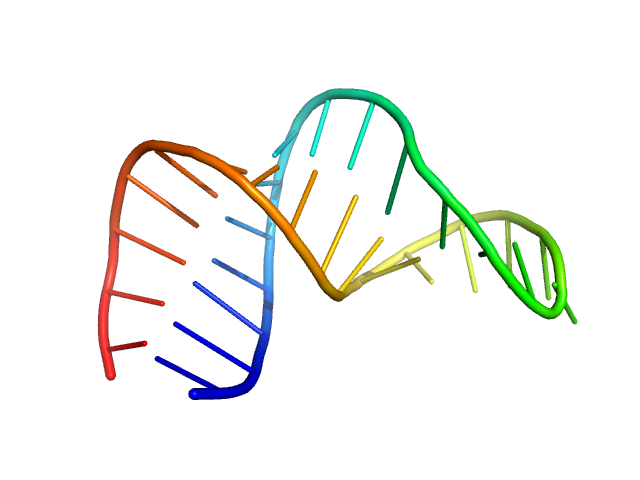
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
|
|
|
|
|
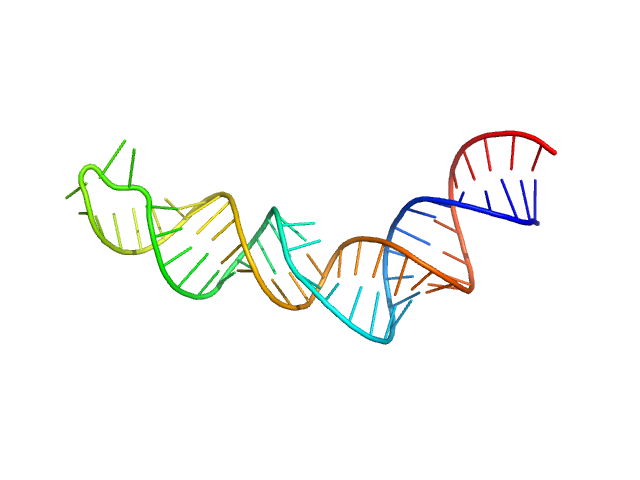
| Sample: |
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
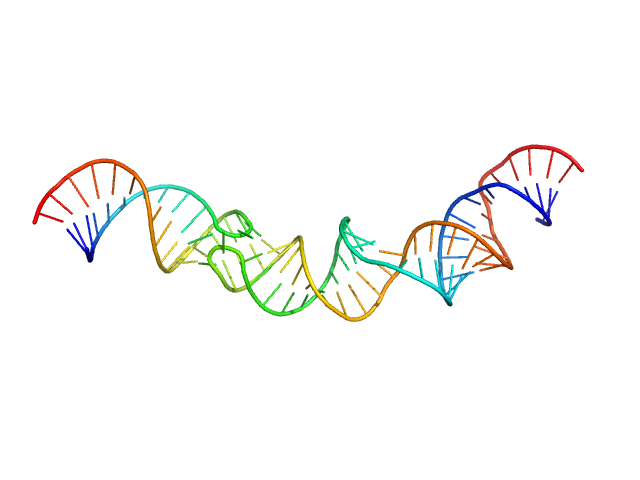
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
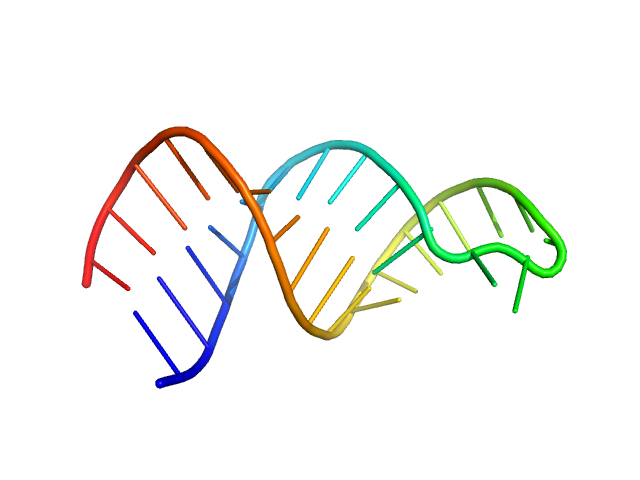
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at CG-3, High Flux Isotope Reactor on 2022 Oct 5
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.3 |
nm |
|
|
|
|
|
|
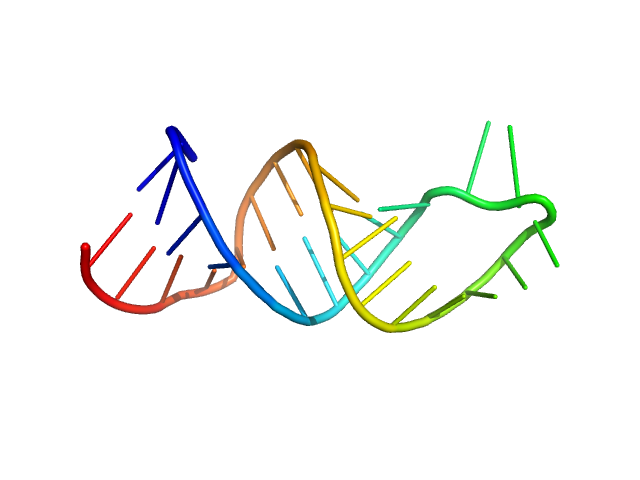
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at CG-3, High Flux Isotope Reactor on 2023 Sep 15
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
|
|
|
|
|
|
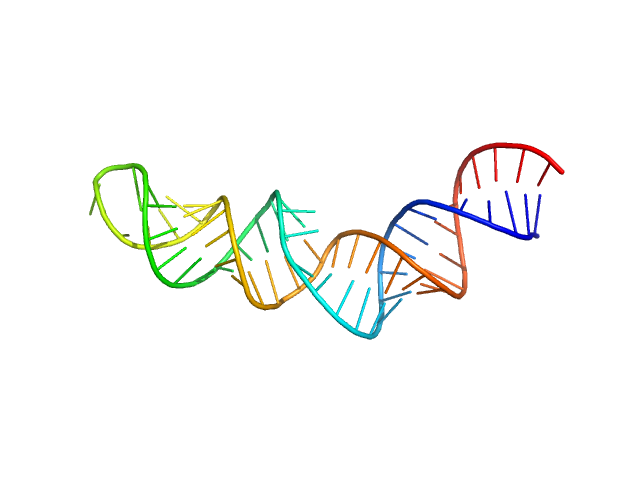
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at CG-3, High Flux Isotope Reactor on 2023 Sep 15
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Optimization of Structure‐Guided Development of Chemical Probes for the Pseudoknot RNA of the Frameshift Element in SARS‐CoV‐2
Angewandte Chemie International Edition (2025)
Ceylan B, Adam J, Toews S, Kaiser F, Dörr J, Scheppa D, Tants J, Smart A, Schoth J, Philipp S, Stirnal E, Ferner J, Richter C, Sreeramulu S, Caliskan N, Schlundt A, Weigand J, Göbel M, Wacker A, Schwalbe H
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Optimization of Structure‐Guided Development of Chemical Probes for the Pseudoknot RNA of the Frameshift Element in SARS‐CoV‐2
Angewandte Chemie International Edition (2025)
Ceylan B, Adam J, Toews S, Kaiser F, Dörr J, Scheppa D, Tants J, Smart A, Schoth J, Philipp S, Stirnal E, Ferner J, Richter C, Sreeramulu S, Caliskan N, Schlundt A, Weigand J, Göbel M, Wacker A, Schwalbe H
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Optimization of Structure‐Guided Development of Chemical Probes for the Pseudoknot RNA of the Frameshift Element in SARS‐CoV‐2
Angewandte Chemie International Edition (2025)
Ceylan B, Adam J, Toews S, Kaiser F, Dörr J, Scheppa D, Tants J, Smart A, Schoth J, Philipp S, Stirnal E, Ferner J, Richter C, Sreeramulu S, Caliskan N, Schlundt A, Weigand J, Göbel M, Wacker A, Schwalbe H
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|