|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 2.5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
The conformational space of RNase P RNA in solution
Nature (2024)
Lee Y, Degenhardt M, Skeparnias I, Degenhardt H, Bhandari Y, Yu P, Stagno J, Fan L, Zhang J, Wang Y
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
306 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 3 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
The conformational space of RNase P RNA in solution
Nature (2024)
Lee Y, Degenhardt M, Skeparnias I, Degenhardt H, Bhandari Y, Yu P, Stagno J, Fan L, Zhang J, Wang Y
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
301 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
The conformational space of RNase P RNA in solution
Nature (2024)
Lee Y, Degenhardt M, Skeparnias I, Degenhardt H, Bhandari Y, Yu P, Stagno J, Fan L, Zhang J, Wang Y
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 8.0 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
The conformational space of RNase P RNA in solution
Nature (2024)
Lee Y, Degenhardt M, Skeparnias I, Degenhardt H, Bhandari Y, Yu P, Stagno J, Fan L, Zhang J, Wang Y
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl,10.0 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
The conformational space of RNase P RNA in solution
Nature (2024)
Lee Y, Degenhardt M, Skeparnias I, Degenhardt H, Bhandari Y, Yu P, Stagno J, Fan L, Zhang J, Wang Y
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
285 |
nm3 |
|
|
|
|
|
|
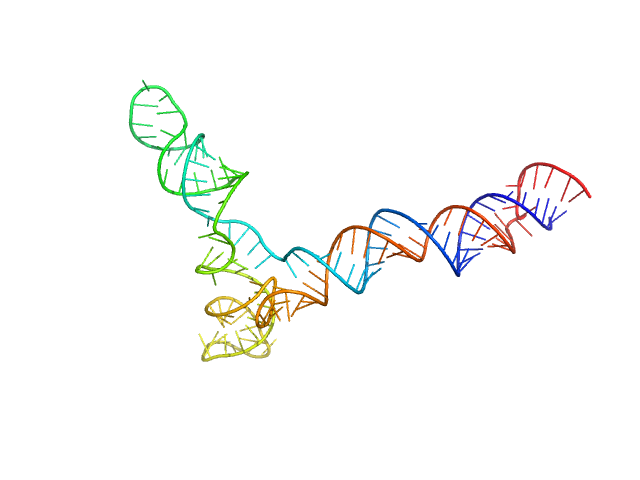
|
| Sample: |
Full stem-loop 5 of SARS-CoV-2 5'genomic end monomer, 48 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sub-element stem-loop 5a from SARS-CoV-2 5'genomic end monomer, 11 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
|
|
|
|
|
|
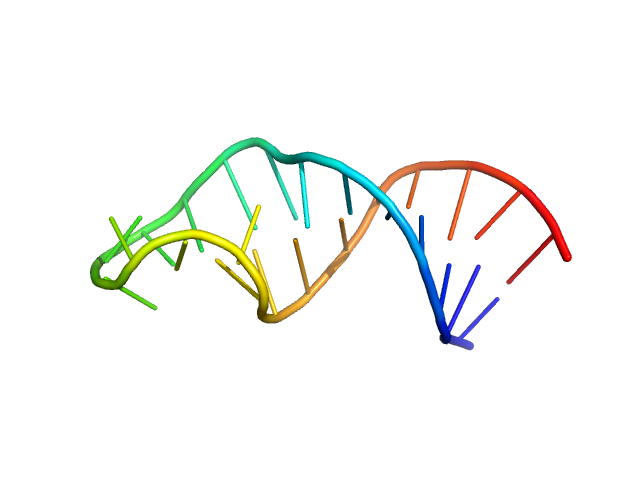
|
| Sample: |
Sub-element stem-loop 5b from SARS-CoV-2 5'genomic end monomer, 8 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Feb 3
|
Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J Am Chem Soc 146(44):30139-30154 (2024)
Mertinkus KR, Oxenfarth A, Richter C, Wacker A, Mata CP, Carazo JM, Schlundt A, Schwalbe H
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
G-quadrupex monomer, 6 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 15
|
G-quadruplex from HBV genome
Trushar Patel
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
G-quadruplex mutant monomer, 6 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 15
|
G-quadruplex from HBV genome
Trushar Patel
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
11 |
nm3 |
|
|