|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGB2_fit1_model1.png)
|
| Sample: |
1,2-dipalmitoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGC2_fit1_model1.png)
|
| Sample: |
1,2-dipalmitoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
|
| Sample: |
Mock recombinant baculovirus, 0 kDa unidentified baculovirus
|
| Buffer: |
TBS (20 mM Tris, 150 mM NaCl),, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 3
|
Rapid screening of in cellulo
grown protein crystals via a small-angle X-ray scattering/X-ray powder diffraction synergistic approach
Journal of Applied Crystallography 53(5) (2020)
Lahey-Rudolph J, Schönherr R, Jeffries C, Blanchet C, Boger J, Ferreira Ramos A, Riekehr W, Triandafillidis D, Valmas A, Margiolaki I, Svergun D, Redecke L
|
|
|
|
|
|
|
|
| Sample: |
High Five insect cells, 0 kDa Trichoplusia ni
|
| Buffer: |
TBS (20 mM Tris, 150 mM NaCl),, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 3
|
Rapid screening of in cellulo
grown protein crystals via a small-angle X-ray scattering/X-ray powder diffraction synergistic approach
Journal of Applied Crystallography 53(5) (2020)
Lahey-Rudolph J, Schönherr R, Jeffries C, Blanchet C, Boger J, Ferreira Ramos A, Riekehr W, Triandafillidis D, Valmas A, Margiolaki I, Svergun D, Redecke L
|
|
|
|
|
|
|
|
| Sample: |
Fe3O4 nanoparticles; radius 5.6 nm (AFM based) monomer, 1 kDa
|
| Buffer: |
water, HCLO4, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 5
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
11.0 |
nm |
| Dmax |
20.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Metaphase chromosomes in 17 mM magnesium chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, 17 mM magnesium chloride, 20 mM sodium chloride, 120 mM potassium chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 May 23
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|
|
|
|
|
|
| Sample: |
Human metaphase chromosomes in 5 mM magnesium chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, 5 mM magnesium chloride, 10 mM sodium chloride, 120 mM potassium chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 Jun 6
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|
|
|
|
|
|
| Sample: |
Human metaphase chromosomes in 1 mM hexaamminecobalt(III) chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, hexaamminecobalt(III) chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 Sep 12
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|
|
|
|
|
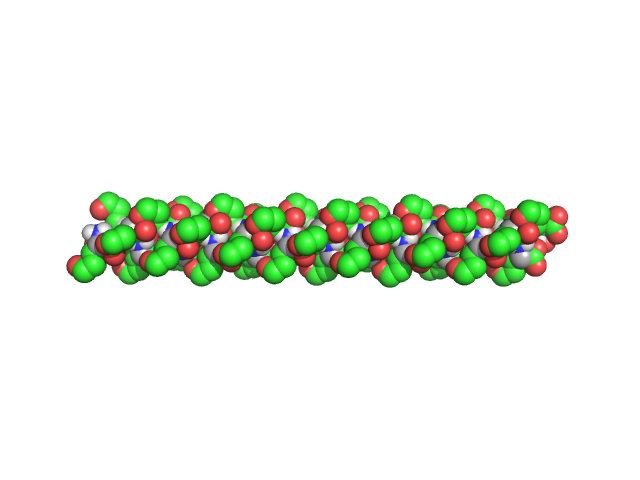
|
| Sample: |
Poly-L-Glutamic Acid, 6 kDa
|
| Buffer: |
Dimethyl Sulfoxide (DMSO), pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 11
|
β2-Type Amyloidlike Fibrils of Poly-l-glutamic Acid Convert into Long, Highly Ordered Helices upon Dissolution in Dimethyl Sulfoxide.
J Phys Chem B 122(50):11895-11905 (2018)
Berbeć S, Dec R, Molodenskiy D, Wielgus-Kutrowska B, Johannessen C, Hernik-Magoń A, Tobias F, Bzowska A, Ścibisz G, Keiderling TA, Svergun D, Dzwolak W
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMX8_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N2) (30% of 9 kDa PEG tails) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|