|
|
|
|
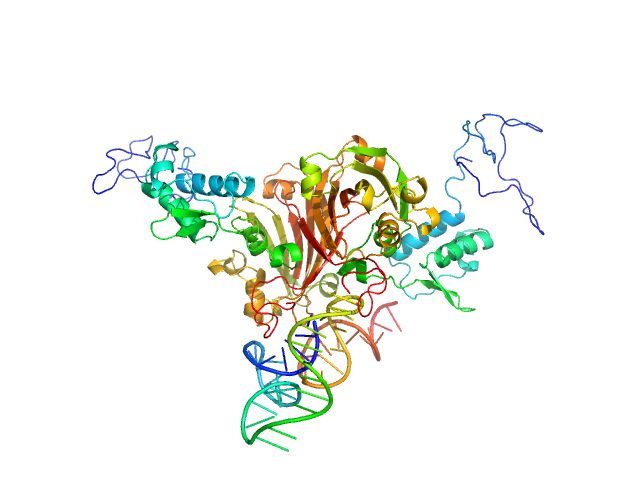
|
| Sample: |
Complex of Rv0792c and Rv0792c_1 monomer, 74 kDa
|
| Buffer: |
25 mM HEPES Buffer; 400 mM NaCl,, pH: 7.2 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 31
|
Structural and Functional Characterization of Rv0792c from Mycobacterium tuberculosis: Identifying Small Molecule Inhibitor against HutC Protein.
Microbiol Spectr :e0197322 (2022)
Chauhan NK, Anand A, Sharma A, Dhiman K, Gosain TP, Singh P, Singh P, Khan E, Chattopadhyay G, Kumar A, Sharma D, Ashish, Sharma TK, Singh R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
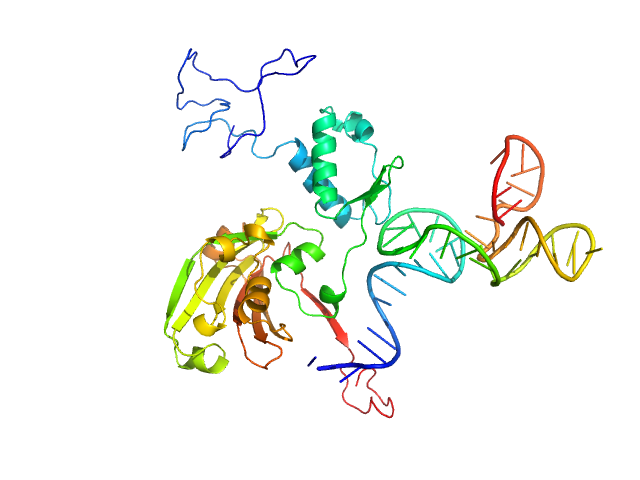
|
| Sample: |
Complex of Rv0792c and Rv0792c_2 monomer, 46 kDa
|
| Buffer: |
25 mM HEPES Buffer; 400 mM NaCl,, pH: 7.2 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 31
|
Structural and Functional Characterization of Rv0792c from Mycobacterium tuberculosis: Identifying Small Molecule Inhibitor against HutC Protein.
Microbiol Spectr :e0197322 (2022)
Chauhan NK, Anand A, Sharma A, Dhiman K, Gosain TP, Singh P, Singh P, Khan E, Chattopadhyay G, Kumar A, Sharma D, Ashish, Sharma TK, Singh R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.8 |
nm |
|
|
|
|
|
|
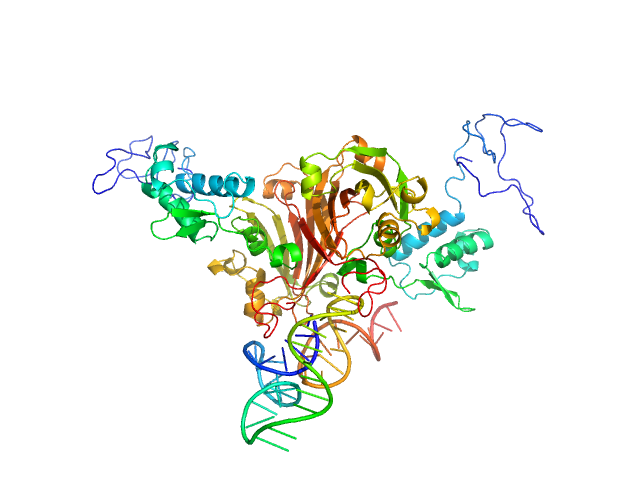
|
| Sample: |
Complex of GntR protein: Aptamer5 monomer, 78 kDa
|
| Buffer: |
25 mM HEPES Buffer; 400 mM NaCl,, pH: 7.2 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 31
|
Structural and Functional Characterization of Rv0792c from Mycobacterium tuberculosis: Identifying Small Molecule Inhibitor against HutC Protein.
Microbiol Spectr :e0197322 (2022)
Chauhan NK, Anand A, Sharma A, Dhiman K, Gosain TP, Singh P, Singh P, Khan E, Chattopadhyay G, Kumar A, Sharma D, Ashish, Sharma TK, Singh R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
DHPC - 1,2-dihexanoyl-sn-glycero-3-phosphocholine None, lipid
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 1
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
Cholate - 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
CHAPS - 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
CHAPS - 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate None, lipid
DMPG - 1,2-dimyristoyl-sn-glycero-3-phospho-sn-glycerol None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
Cholate - 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid None, lipid
DMPG - 1,2-dimyristoyl-sn-glycero-3-phospho-sn-glycerol None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DHPC - 1,2-dihexanoyl-sn-glycero-3-phosphocholine None, lipid
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
DMPG - 1,2-dimyristoyl-sn-glycero-3-phospho-sn-glycerol None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
CHAPS - 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate None, lipid
POPC - 2-oleoyl-1-palmitoyl-sn-glyecro-3-phosphocholine None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|