|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab (Non-structural protein 10 - Δ4386-4392) monomer, 14 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM HEPES, 300 mM NaCl, 1.5% (v/v) glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Nov 25
|
Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering
International Journal of Molecular Sciences 24(17):13649 (2023)
Knecht W, Fisher S, Lou J, Sele C, Ma S, Rasmussen A, Pinotsis N, Kozielski F
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab (Non-structural protein 10 - Δ4254-4262; Δ4386-4392) monomer, 13 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM HEPES, 300 mM NaCl, 1.5% (v/v) glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Nov 25
|
Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering
International Journal of Molecular Sciences 24(17):13649 (2023)
Knecht W, Fisher S, Lou J, Sele C, Ma S, Rasmussen A, Pinotsis N, Kozielski F
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab (Non-structural protein 10) monomer, 16 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM HEPES, 300 mM NaCl, 1.5% (v/v) glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 6
|
Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering
International Journal of Molecular Sciences 24(17):13649 (2023)
Knecht W, Fisher S, Lou J, Sele C, Ma S, Rasmussen A, Pinotsis N, Kozielski F
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab (Non-structural protein 10) monomer, 15 kDa Middle East respiratory … protein
|
| Buffer: |
10 mM HEPES, 300 mM NaCl, 1.5% (v/v) glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 6
|
Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering
International Journal of Molecular Sciences 24(17):13649 (2023)
Knecht W, Fisher S, Lou J, Sele C, Ma S, Rasmussen A, Pinotsis N, Kozielski F
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
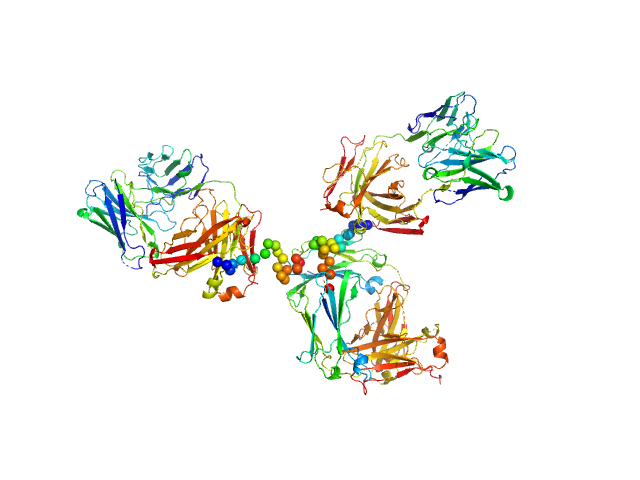
| Sample: |
Palivizumab IgG monomer, 145 kDa Commercial (Synagis) protein
|
| Buffer: |
PBS (137 mM NaCl, 2.7 mM KCl, 12 mM HPO4 2−/H2PO4 −), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
Respiratory syncytial virus (RSV)-approved monoclonal antibody Palivizumab as ligand for anti-idiotype nanobody-based synthetic cytokine receptors
Journal of Biological Chemistry :105270 (2023)
Ettich J, Wittich C, Moll J, Behnke K, Floss D, Reiners J, Christmann A, Lang P, Smits S, Kolmar H, Scheller J
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
|
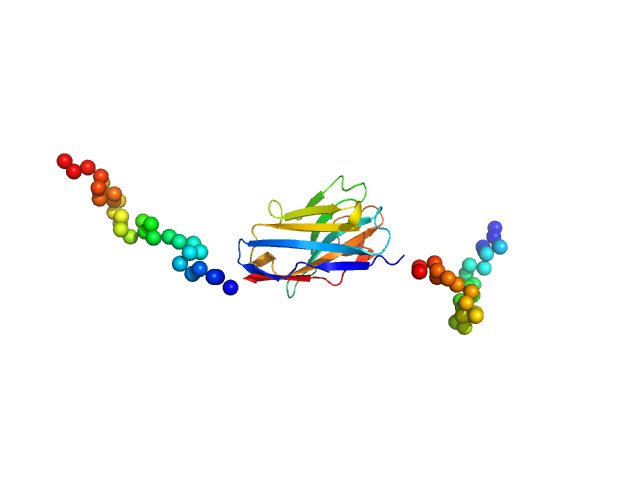
| Sample: |
Anti-Idiotypic-Palivizumab-Nanobody1 monomer, 20 kDa Lama glama protein
|
| Buffer: |
PBS (137 mM NaCl, 2.7 mM KCl, 12 mM HPO4 2−/H2PO4 −), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
Respiratory syncytial virus (RSV)-approved monoclonal antibody Palivizumab as ligand for anti-idiotype nanobody-based synthetic cytokine receptors
Journal of Biological Chemistry :105270 (2023)
Ettich J, Wittich C, Moll J, Behnke K, Floss D, Reiners J, Christmann A, Lang P, Smits S, Kolmar H, Scheller J
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
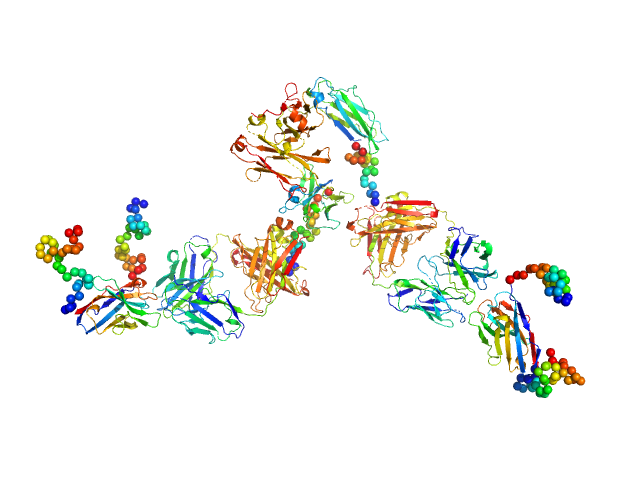
|
| Sample: |
Palivizumab IgG monomer, 145 kDa Commercial (Synagis) protein
Anti-Idiotypic-Palivizumab-Nanobody1 monomer, 20 kDa Lama glama protein
|
| Buffer: |
PBS (137 mM NaCl, 2.7 mM KCl, 12 mM HPO4 2−/H2PO4 −), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
Respiratory syncytial virus (RSV)-approved monoclonal antibody Palivizumab as ligand for anti-idiotype nanobody-based synthetic cytokine receptors
Journal of Biological Chemistry :105270 (2023)
Ettich J, Wittich C, Moll J, Behnke K, Floss D, Reiners J, Christmann A, Lang P, Smits S, Kolmar H, Scheller J
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.4 |
nm |
|
|
|
|
|
|
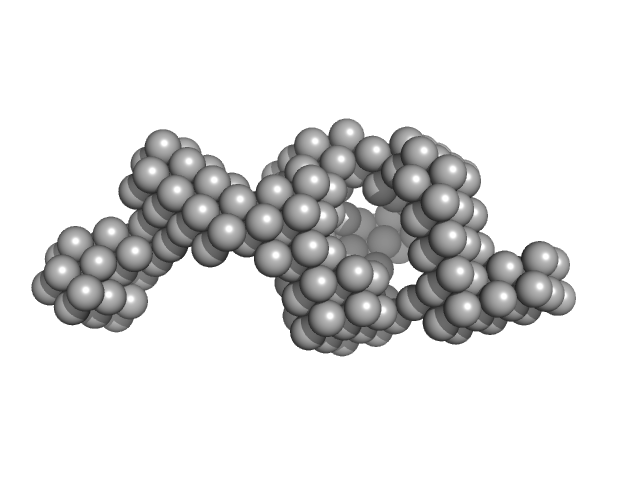
|
| Sample: |
Probable transcriptional regulatory protein (Probably AsnC-family) dimer, 36 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Mar 1
|
Mycobacterium tuberculosis Rv2324 is a multifunctional feast/famine regulatory protein involved in growth, DNA replication and damage control.
Int J Biol Macromol 252:126459 (2023)
Dubey S, Maurya RK, Shree S, Kumar S, Jahan F, Krishnan MY, Ramachandran R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Probable transcriptional regulatory protein (Probably AsnC-family) octamer, 143 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM EDTA, 2 mM methionine, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Dec 5
|
Mycobacterium tuberculosis Rv2324 is a multifunctional feast/famine regulatory protein involved in growth, DNA replication and damage control.
Int J Biol Macromol 252:126459 (2023)
Dubey S, Maurya RK, Shree S, Kumar S, Jahan F, Krishnan MY, Ramachandran R
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
246 |
nm3 |
|
|
|
|
|
|
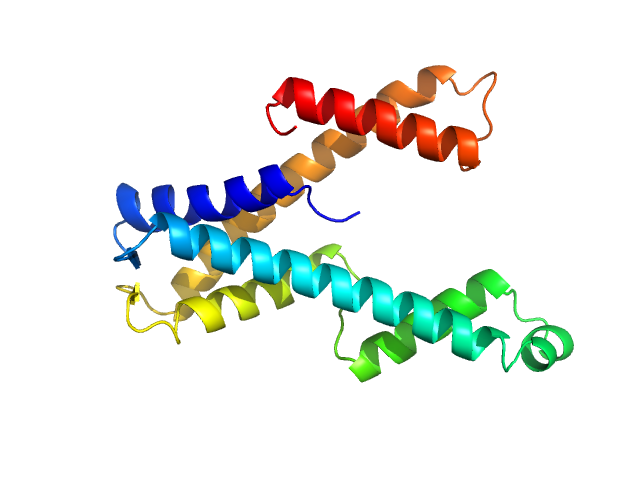
|
| Sample: |
DUF507 family protein monomer, 22 kDa Aquifex aeolicus (strain … protein
|
| Buffer: |
20 mM Hepes pH 7.2, 400 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 7
|
Crystal structure of domain of unknown function 507 (DUF507) reveals a new protein fold.
Sci Rep 13(1):13496 (2023)
McKay CE, Cheng J, Tanner JJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|