|
|
|
|
|
| Sample: |
Human dystrophin central domain R16-24 fragment monomer, 127 kDa protein
|
| Buffer: |
NaP 20 mM, NaCl 300 mM, EDTA 1 mM, Glycérol 2%, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 17
|
Dystrophin SAXS data
Raphael Dos Santos Morais
|
| RgGuinier |
9.3 |
nm |
| Dmax |
34.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Human dystrophin central domain R16-24 del45-55 fragment monomer, 58 kDa protein
|
| Buffer: |
NaP 20 mM, NaCl 300 mM, EDTA 1 mM, Glycérol 2%, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Sep 28
|
Dystrophin SAXS data
Raphael Dos Santos Morais
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
Human dystrophin central domain R16-24 del45-51 fragment monomer, 85 kDa protein
|
| Buffer: |
NaP 20 mM, NaCl 300 mM, EDTA 1 mM, Glycérol 2%, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Jul 13
|
Dystrophin SAXS data
Raphael Dos Santos Morais
|
| RgGuinier |
7.0 |
nm |
| Dmax |
26.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Human dystrophin central domain R16-24 del44-54 fragment monomer, 57 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 300 mM NaCl, 1 mM EDTA, 2% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 8
|
Dystrophin SAXS data
Raphael Dos Santos Morais
|
| RgGuinier |
6.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
|
|
|
|
|
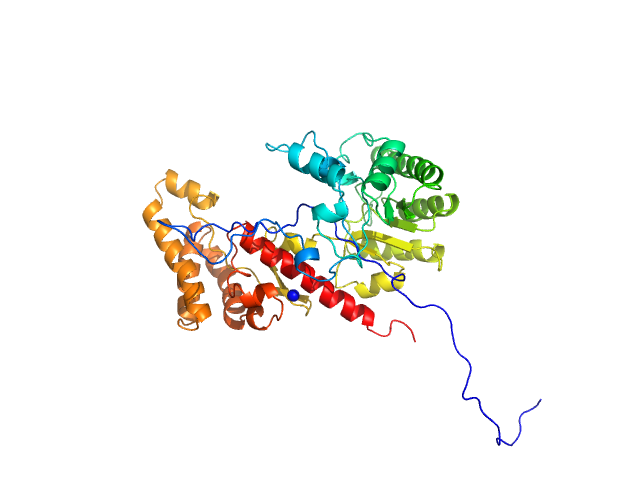
| Sample: |
Cell division protein FtsQ monomer, 11 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM phosphate buffer, 25 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at 5-ID-D DND-CAT, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jun 28
|
An Arg/Ala-rich helix in the N-terminal region of M. tuberculosis FtsQ is a potential membrane anchor of the Z-ring.
Commun Biol 6(1):311 (2023)
Smrt ST, Escobar CA, Dey S, Cross TA, Zhou HX
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
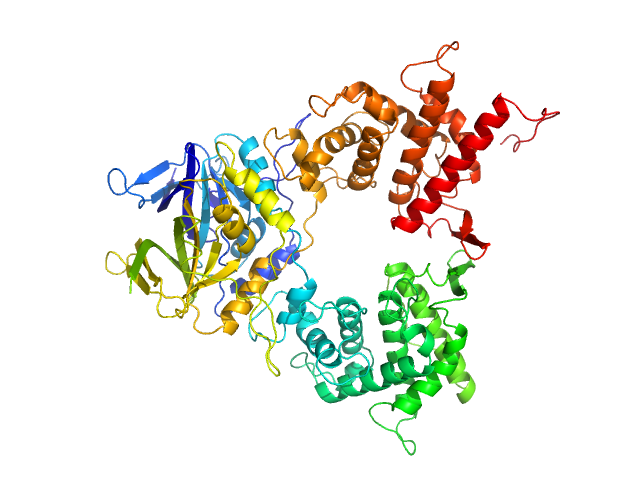
| Sample: |
DNA-guanine transglycosylase - D95A mutant monomer, 50 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
100 mM KCl, 50 mM Tris pH 7.0, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 22
|
7-Deazaguanines in DNA: functional and structural elucidation of a DNA modification system.
Nucleic Acids Res (2023)
Gedara SH, Wood E, Gustafson A, Liang C, Hung SH, Savage J, Phan P, Luthra A, de Crécy-Lagard V, Dedon P, Swairjo MA, Iwata-Reuyl D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DGQHR domain-containing protein dimer, 88 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
100 mM KCl, 50 mM Tris pH 7.0, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 22
|
7-Deazaguanines in DNA: functional and structural elucidation of a DNA modification system.
Nucleic Acids Res (2023)
Gedara SH, Wood E, Gustafson A, Liang C, Hung SH, Savage J, Phan P, Luthra A, de Crécy-Lagard V, Dedon P, Swairjo MA, Iwata-Reuyl D
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 31 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Mar 18
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|

|
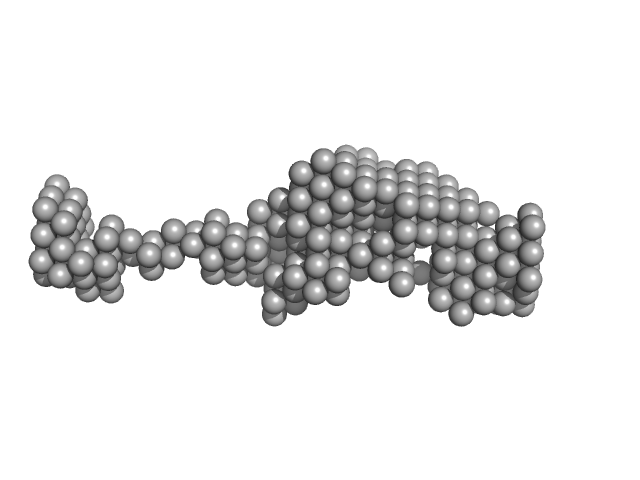
| Sample: |
Cell wall assembly regulator SMI1 monomer, 40 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 4
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 57 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Mar 7
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|