|
|
|
|
|
| Sample: |
Human immunoglobulin SAP1.3 - kappa chain dimer, 48 kDa Homo sapiens protein
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain dimer, 99 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain C226S dimer, 99 kDa Homo sapiens protein
Human immunoglobulin SAP1.3 - kappa chain C214S dimer, 48 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human immunoglobulin SAP1.3 - kappa chain C214S dimer, 48 kDa Homo sapiens protein
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain C227S dimer, 99 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human immunoglobulin SAP1.3 - kappa chain dimer, 48 kDa Homo sapiens protein
Human immunoglobulin gamma 2 (IgG2) SAP1.3 - heavy chain C226S/C227S dimer, 99 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Sep 28
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDVV6_fit1_model1.png)
|
| Sample: |
Drebrin monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
17 mM NaH2PO4, 3 mM Na2HPO4, 50 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 7
|
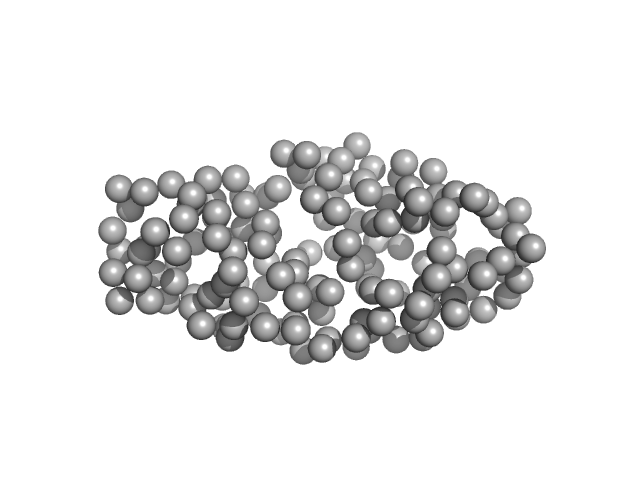
Dynamic Interchange of Local Residue-Residue Interactions in the Largely Extended Single Alpha-Helix in Drebrin
Biochemical Journal (2025)
Varga S, Péterfia B, Dudola D, Farkas V, Jeffries C, Permi P, Gáspári Z
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Apr 20
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
20464 |
nm3 |
|
|
|
|
|
|

|
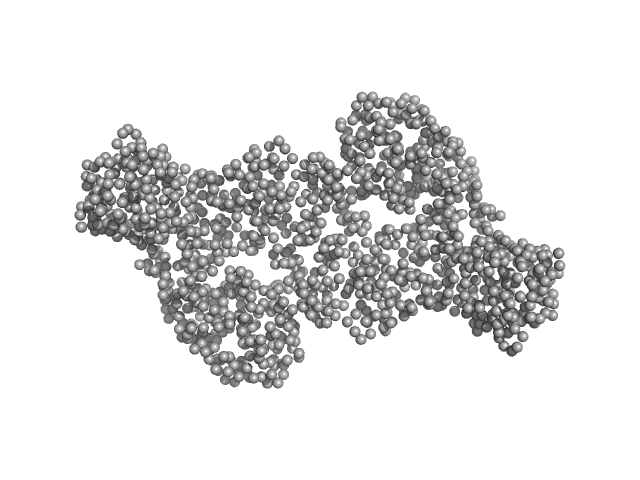
| Sample: |
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
271263 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Histone H3.3 dimer, 8 kDa Homo sapiens protein
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Histone H3.3 monomer, 4 kDa Homo sa protein
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Sep 17
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
DNA methyltransferase 3 beta (413-853) dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Nov 2
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
191 |
nm3 |
|
|