|
|
|
|
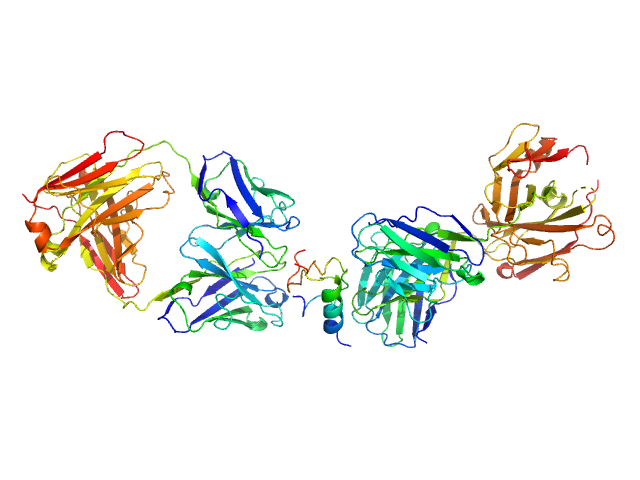
|
| Sample: |
HUI-018 Fab monomer, 47 kDa Mus musculus protein
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Sus scrofa protein
OXI-005 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Novo Nordisk A/S on 2019 Mar 28
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
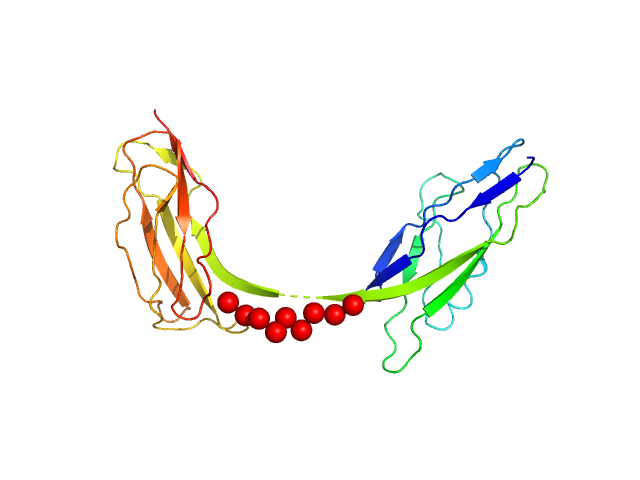
|
| Sample: |
Intimin, D00-D0 domain (6xHis tagged) monomer, 23 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
10 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 12
|
The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep 10(1):21249 (2020)
Weikum J, Kulakova A, Tesei G, Yoshimoto S, Jægerum LV, Schütz M, Hori K, Skepö M, Harris P, Leo JC, Morth JP
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Intimin (D0-D1 domain, 6xHis tagged) monomer, 23 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
10 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 12
|
The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep 10(1):21249 (2020)
Weikum J, Kulakova A, Tesei G, Yoshimoto S, Jægerum LV, Schütz M, Hori K, Skepö M, Harris P, Leo JC, Morth JP
|
| RgGuinier |
2.2 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
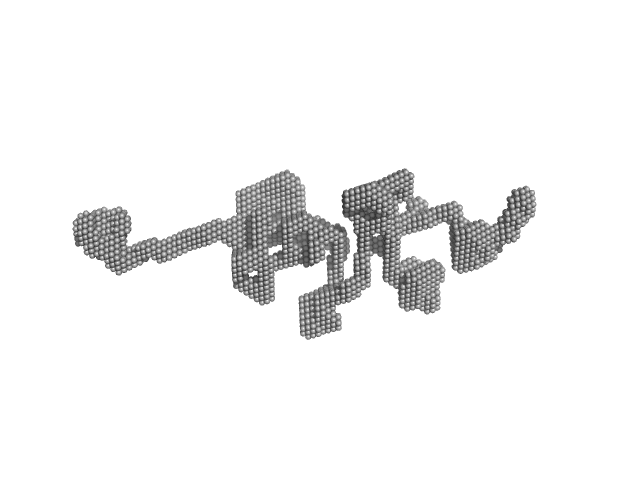
|
| Sample: |
Nucleolar RNA helicase 2 dimer, 182 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 25
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
7.0 |
nm |
| Dmax |
31.9 |
nm |
| VolumePorod |
681 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-783 dimer, 138 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-710 dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
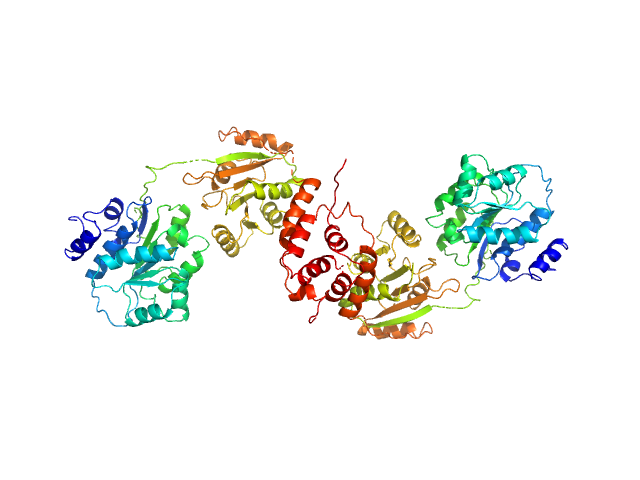
|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-620 dimer, 98 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolar RNA helicase 2 monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 3
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
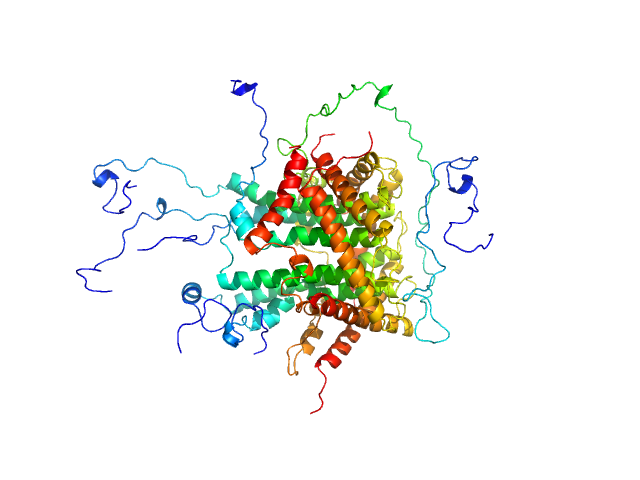
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM8 trimer, 29 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM13 trimer, 34 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM23 monomer, 24 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Apr 24
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.3 |
nm |
|
|
|
|
|
|
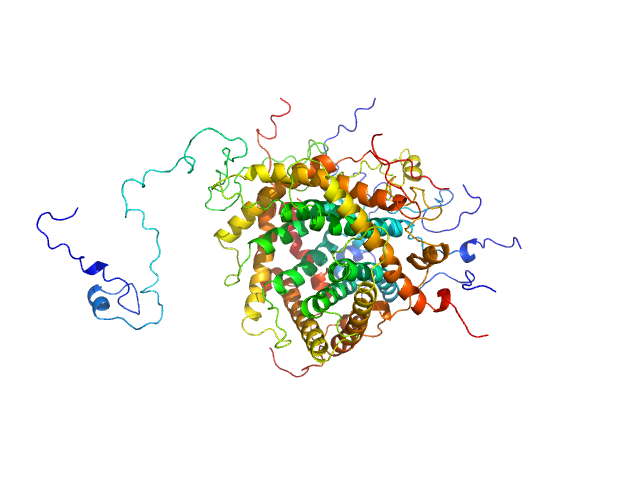
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM9 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM23 monomer, 24 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 18
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
146 |
nm3 |
|
|