|
|
|
|
|
| Sample: |
Ammonium sensor/transducer trimer, 227 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, 0.09%(w/v) CYMAL-5, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
482 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 5
|
Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res 46(1):387-402 (2018)
Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov AV, Luisi BF
|
| RgGuinier |
5.3 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
ATP-dependent RNA helicase RhlB monomer, 47 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Feb 11
|
Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res 46(1):387-402 (2018)
Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov AV, Luisi BF
|
| RgGuinier |
5.4 |
nm |
| Dmax |
29.5 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
ATP-dependent RNA helicase RhlB monomer, 47 kDa Escherichia coli protein
Enolase dimer, 91 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Jul 16
|
Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res 46(1):387-402 (2018)
Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov AV, Luisi BF
|
| RgGuinier |
6.4 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytohesin-3 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 13
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cytohesin-3 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.001 mM inositol 1,3,4,5-tetrakisphosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Mar 21
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Grp1 63-399 E161A 6GS Arf6 Q67L fusion protein monomer, 61 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Grp1 63-399 E161A 6GS Arf6 Q67L His fusion protein monomer, 62 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
|
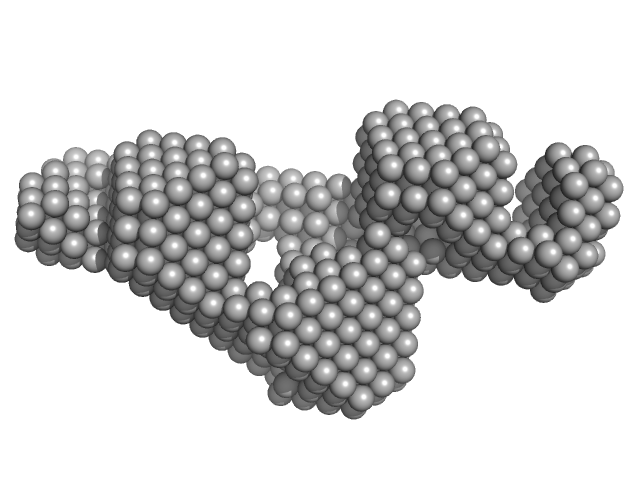
| Sample: |
Grp1 63-399 E161A 6GS Arf6 Q67L SUMO fusion protein monomer, 72 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Grp1 63-399 E161A Arf6 Q67L fusion protein monomer, 60 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
86 |
nm3 |
|
|