|
|
|
|
|
| Sample: |
Bacteriophage phi-X174 monomer, 0 kDa protein
|
| Buffer: |
0.06 M NH4Cl2, 0.09 M NaCl, 0.1 M KCl, 1 mM MgS04, 1 mM CaCl2, 0.1 M Tris-HCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Structural changes of tailless bacteriophage ΦX174 during penetration of bacterial cell walls.
Proc Natl Acad Sci U S A 114(52):13708-13713 (2017)
Sun Y, Roznowski AP, Tokuda JM, Klose T, Mauney A, Pollack L, Fane BA, Rossmann MG
|
|
|
|
|
|
|
|
| Sample: |
Bacteriophage phi-X174 monomer, 0 kDa protein
|
| Buffer: |
0.15 mg/mL LPS, 0.06 M NH4Cl2, 0.09 M NaCl, 0.1 M KCl, 1 mM MgS04, 1 mM CaCl2, 0.1 M Tris-HCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Structural changes of tailless bacteriophage ΦX174 during penetration of bacterial cell walls.
Proc Natl Acad Sci U S A 114(52):13708-13713 (2017)
Sun Y, Roznowski AP, Tokuda JM, Klose T, Mauney A, Pollack L, Fane BA, Rossmann MG
|
|
|
|
|
|
|
|
| Sample: |
Bacteriophage phi-X174 monomer, 0 kDa protein
|
| Buffer: |
0.15 mg/mL LPS, 0.06 M NH4Cl2, 0.09 M NaCl, 0.1 M KCl, 1 mM MgS04, 1 mM CaCl2, 0.1 M Tris-HCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Structural changes of tailless bacteriophage ΦX174 during penetration of bacterial cell walls.
Proc Natl Acad Sci U S A 114(52):13708-13713 (2017)
Sun Y, Roznowski AP, Tokuda JM, Klose T, Mauney A, Pollack L, Fane BA, Rossmann MG
|
|
|
|
|
|
|
|
| Sample: |
Bacteriophage phi-X174 monomer, 0 kDa protein
|
| Buffer: |
0.15 mg/mL LPS, 0.06 M NH4Cl2, 0.09 M NaCl, 0.1 M KCl, 1 mM MgS04, 1 mM CaCl2, 0.1 M Tris-HCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Structural changes of tailless bacteriophage ΦX174 during penetration of bacterial cell walls.
Proc Natl Acad Sci U S A 114(52):13708-13713 (2017)
Sun Y, Roznowski AP, Tokuda JM, Klose T, Mauney A, Pollack L, Fane BA, Rossmann MG
|
|
|
|
|
|
|

|
| Sample: |
Nucleoplasmin core + A2 pentamer, 81 kDa Xenopus laevis protein
Histone H2A (ΔAla127) pentamer, 69 kDa Xenopus laevis protein
Histone H2B 1.1 (Ser33Thr) pentamer, 67 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Tris. 150 mM NaCl, 1 mM EDTA, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Jan 7
|
Dynamic intramolecular regulation of the histone chaperone nucleoplasmin controls histone binding and release.
Nat Commun 8(1):2215 (2017)
Warren C, Matsui T, Karp JM, Onikubo T, Cahill S, Brenowitz M, Cowburn D, Girvin M, Shechter D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
402 |
nm3 |
|
|
|
|
|
|
|
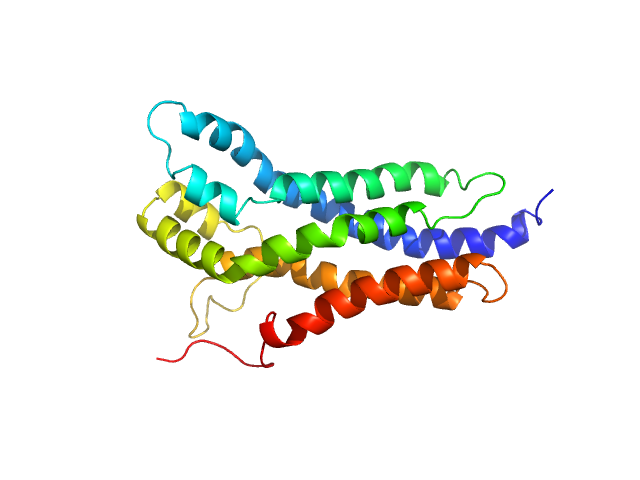
| Sample: |
BCR-ABL p210 fusion protein (DH-PH) monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 13
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
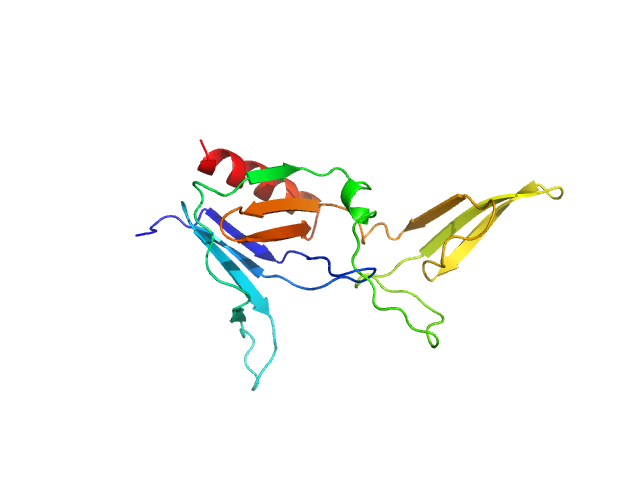
| Sample: |
BCR-ABL p210 fusion protein monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 13
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
BCR-ABL p210 fusion protein (PH domain) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 10
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DHH subfamily 1 protein dimer, 70 kDa Streptococcus pneumoniae serotype … protein
|
| Buffer: |
20mM Tris, 200 mM NaCl, 5%(v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 23
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
T.maritima PDE dimer, 76 kDa Thermotoga maritima protein
|
| Buffer: |
25mM Tris 500mM NaCl 3% (v/v) glycerol 2mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 17
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.8 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
115 |
nm3 |
|
|