|
|
|
|

|
| Sample: |
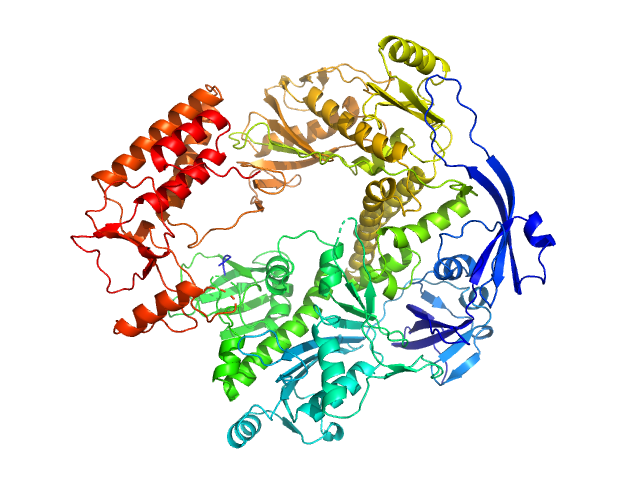
Pomacea maculata perivitellin 1 dodecamer, 278 kDa Pomacea maculata protein
|
| Buffer: |
100 mM Phosphate Buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 26
|
Convergent evolution of plant and animal embryo defences by hyperstable non-digestible storage proteins.
Sci Rep 7(1):15848 (2017)
Pasquevich MY, Dreon MS, Qiu JW, Mu H, Heras H
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
537 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA polymerase E9 monomer, 117 kDa Vaccinia virus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 4 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 27
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA polymerase processivity factor component A20 C-ter fragment monomer, 17 kDa Vaccinia virus protein
|
| Buffer: |
25 mM Tris-HCl, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
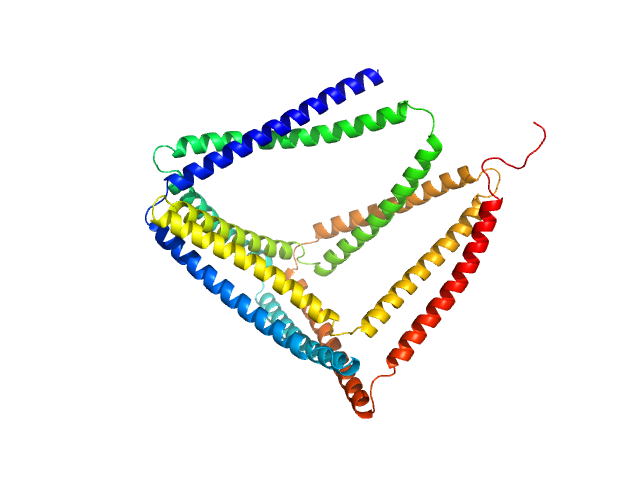
TET12(1.10)SN-f5 monomer, 53 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Oct 28
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
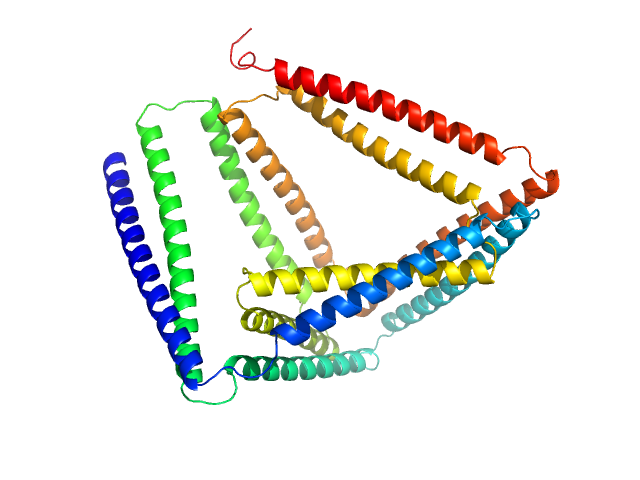
TET12(1.10)SN-c6 monomer, 54 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 10
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
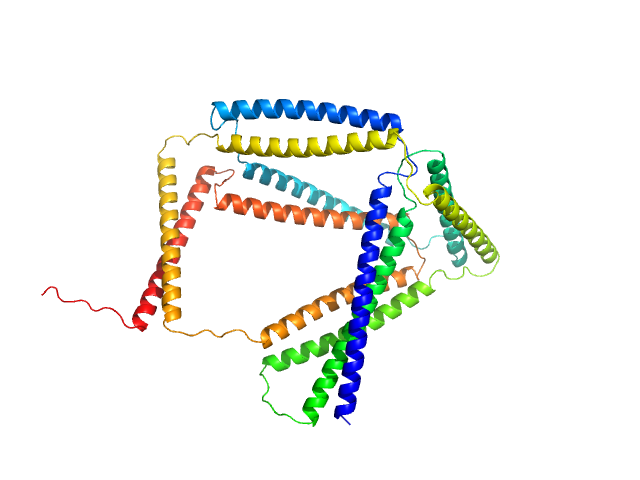
TET12(1.10)SN-f9 monomer, 57 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 10
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
153 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PYR16(4.6)SN-f5 monomer, 72 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 16
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
|
|
|
|

|
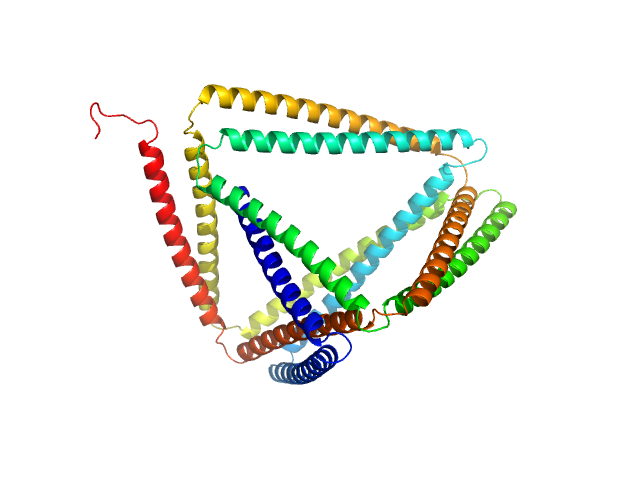
| Sample: |
TRIP18(7.5R)SN-f5 monomer, 82 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 10
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
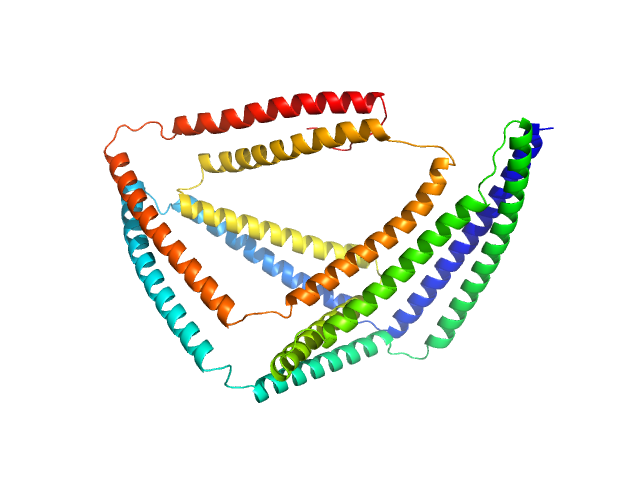
| Sample: |
TET12(2.3)SN-f5b monomer, 57 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 10
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
TET12(1.10)S-f5 monomer, 53 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 7
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
210 |
nm3 |
|
|