|
|
|
|
|
| Sample: |
Monoubiquitinated Rab5 at K165 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2016 Nov 21
|
Site-specific monoubiquitination downregulates Rab5 by disrupting effector binding and guanine nucleotide conversion.
Elife 6 (2017)
Shin D, Na W, Lee JH, Kim G, Baek J, Park SH, Choi CY, Lee S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
Monoubiquitinated Rab5 at K165 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2016 Nov 21
|
Site-specific monoubiquitination downregulates Rab5 by disrupting effector binding and guanine nucleotide conversion.
Elife 6 (2017)
Shin D, Na W, Lee JH, Kim G, Baek J, Park SH, Choi CY, Lee S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
Monoubiquitinated Rab5 at K165 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2016 Nov 21
|
Site-specific monoubiquitination downregulates Rab5 by disrupting effector binding and guanine nucleotide conversion.
Elife 6 (2017)
Shin D, Na W, Lee JH, Kim G, Baek J, Park SH, Choi CY, Lee S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
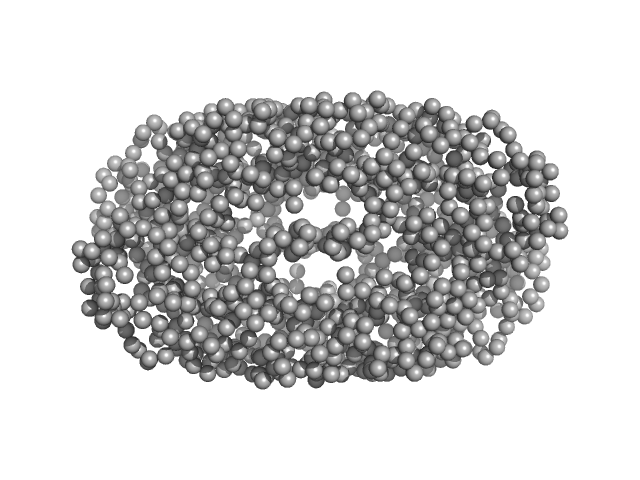
Geobacillus stearothermophilus DnaB1-300 tetramer, 138 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl and 5 mM β-ME, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 Oct 18
|
Structural analyses of the bacterial primosomal protein DnaB reveal that it is a tetramer and forms a complex with a primosomal re-initiation protein.
J Biol Chem 292(38):15744-15757 (2017)
Li YC, Naveen V, Lin MG, Hsiao CD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
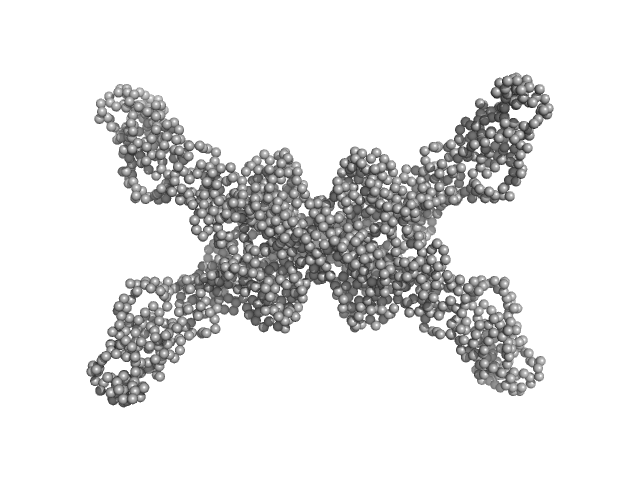
Geobacillus stearothermophilus DnaB full-length tetramer, 214 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl and 5 mM β-ME, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 Mar 12
|
Structural analyses of the bacterial primosomal protein DnaB reveal that it is a tetramer and forms a complex with a primosomal re-initiation protein.
J Biol Chem 292(38):15744-15757 (2017)
Li YC, Naveen V, Lin MG, Hsiao CD
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
432 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidylprolyl isomerase monomer, 54 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM KCl, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Structural and molecular comparison of bacterial and eukaryotic trigger factors.
Sci Rep 7(1):10680 (2017)
Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trigger factor-like protein TIG, Chloroplastic monomer, 53 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM KCl, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Structural and molecular comparison of bacterial and eukaryotic trigger factors.
Sci Rep 7(1):10680 (2017)
Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family). dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Archaeal biofilm regulator 1 (AbfR1) mutant Y84E S87D phosphomimic mutant dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
229 |
nm3 |
|
|