|
|
|
|
|
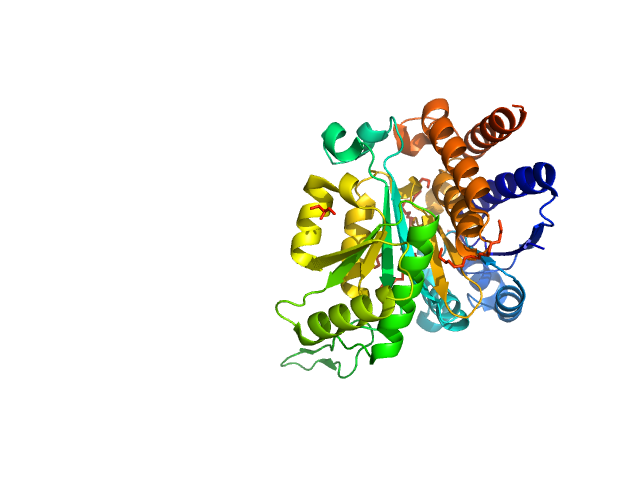
| Sample: |
3-isopropylmalate dehydrogenase dimer, 73 kDa Thermus thermophilus protein
|
| Buffer: |
25 mM MOPS/NaOH, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Nov 16
|
Dual Role of the Active Site Residues of Thermus thermophilus 3-Isopropylmalate Dehydrogenase: Chemical Catalysis and Domain Closure.
Biochemistry 55(3):560-74 (2016)
Gráczer É, Szimler T, Garamszegi A, Konarev PV, Lábas A, Oláh J, Palló A, Svergun DI, Merli A, Závodszky P, Weiss MS, Vas M
|
|
|
|
|
|
|
|
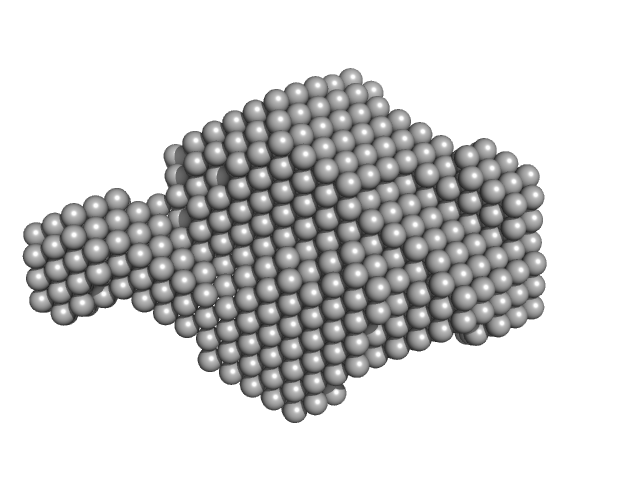
| Sample: |
Aureochrome 1a (N-terminally truncated) dimer, 53 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
20 mM HEPES 100 mM NaCl 10 mM MgCl2 5% w/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2015 Mar 11
|
Blue light-induced LOV domain dimerization enhances the affinity of Aureochrome 1a for its target DNA sequence.
Elife 5:e11860 (2016)
Heintz U, Schlichting I
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a-A´α-LOV-Jα dimer, 36 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 16
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a-A´α-LOV-Jα dimer, 36 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 16
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
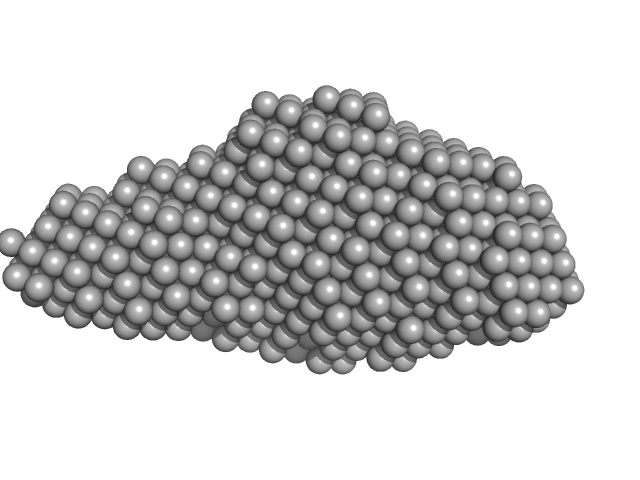
| Sample: |
Aureochrome1a- A´α-LOV-Jα (Dark State) dimer, 32 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 9
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a- A´α-LOV-Jα (Light State) dimer, 32 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 9
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cardiac myosin binding protein-C: domains C5-C6-C7 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 250 mM NaCl, 2 mM TCEP, 0.02% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 18
|
Clinically Linked Mutations in the Central Domains of Cardiac Myosin-Binding Protein C with Distinct Phenotypes Show Differential Structural Effects.
Structure 24(1):105-115 (2016)
Nadvi NA, Michie KA, Kwan AH, Guss JM, Trewhella J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
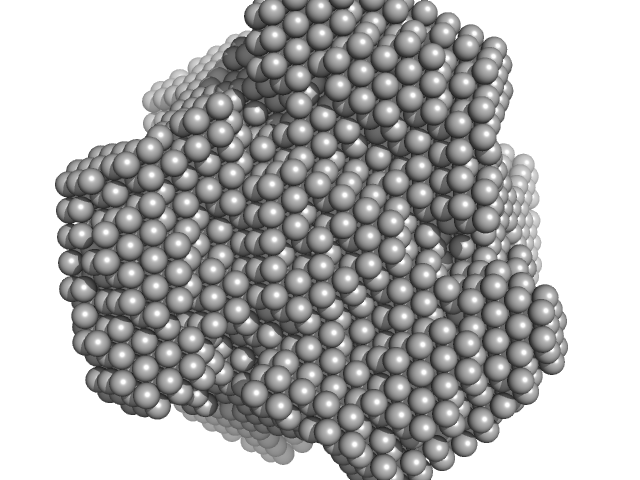
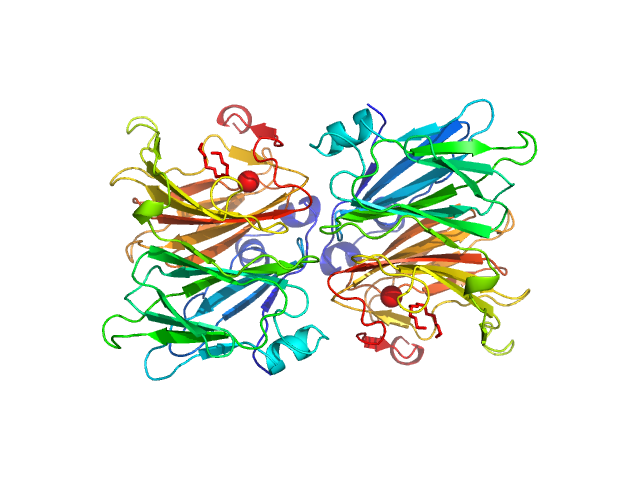
| Sample: |
Inorganic pyrophosphatase (PPase) from E. coli hexamer, 117 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 30
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
|
|
|
|
|
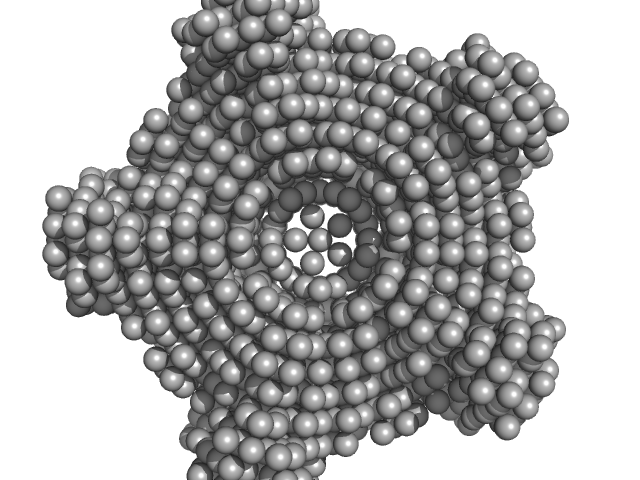
| Sample: |
Class I fructose-1,6-bisphosphate aldolase (FbaB) from E. coli decamer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 30
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
484 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
5-keto-4-deoxyuronate isomerase (KduI) from E. coli None, Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 20
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
|
|