|
|
|
|
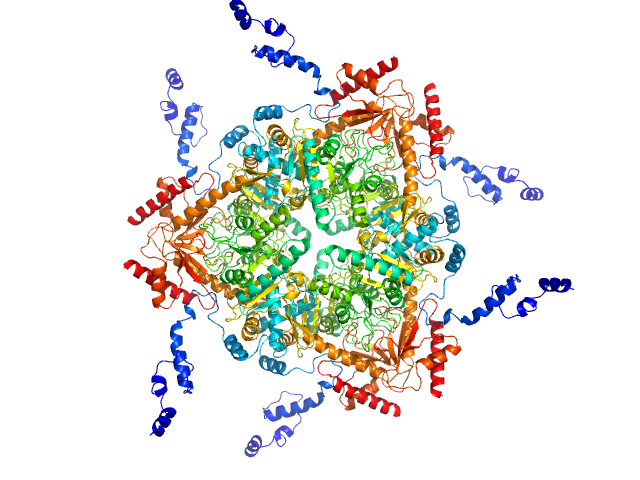
|
| Sample: |
Glutamate decarboxylase alpha (GadA) from E. coli monomer, 53 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 20
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.8 |
nm |
| VolumePorod |
410 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human Calumenin monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
25 mM Na-HEPES, 25 mM NaCl, 2.5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Feb 12
|
Ca-Dependent Folding of Human Calumenin.
PLoS One 11(3):e0151547 (2016)
Mazzorana M, Hussain R, Sorensen T
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
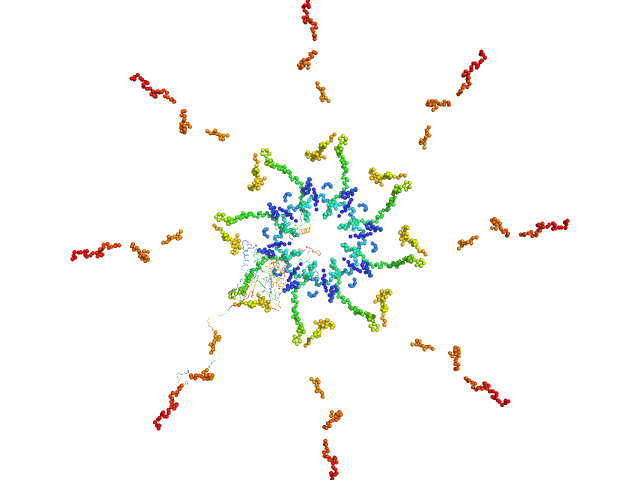
|
| Sample: |
Callose synthase octamer, 633 kDa Arabidopsis thaliana protein
|
| Buffer: |
Tris, 50 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Structural Characterization of Cell Wall and Plasma Membrane Proteins of Arabidopsis thaliana
University of Hamburg Dissertation 8022 (2016)
Haifa El Kilani
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
1033 |
nm3 |
|
|
|
|
|
|
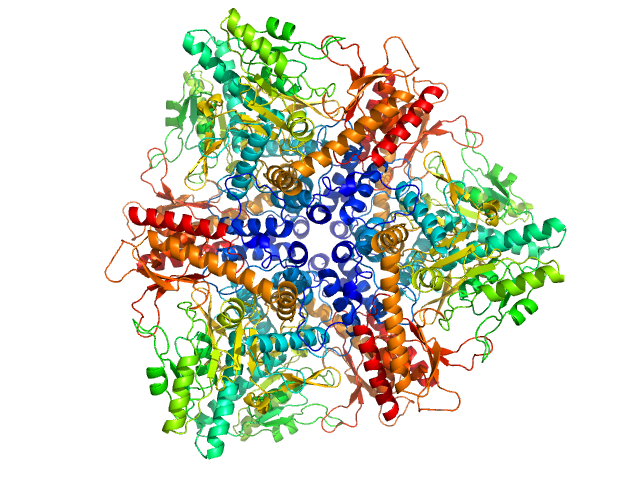
|
| Sample: |
Glutamate decarboxylase alpha (GadA) from E. coli monomer, 53 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 29
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.4 |
nm |
| VolumePorod |
450 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
FI10506p monomer, 23 kDa Drosophila melanogaster protein
|
| Buffer: |
20mM Tris/HCl 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 21
|
Intrinsic Disorder of the C-Terminal Domain of Drosophila Methoprene-Tolerant Protein.
PLoS One 11(9):e0162950 (2016)
Kolonko M, Ożga K, Hołubowicz R, Taube M, Kozak M, Ożyhar A, Greb-Markiewicz B
|
| RgGuinier |
5.1 |
nm |
| Dmax |
22.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Latency-associated nuclear antigen tetramer, 87 kDa Murid herpesvirus 4 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
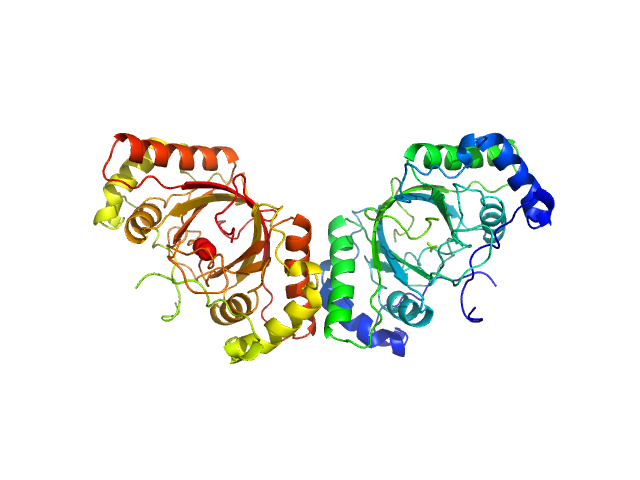
|
| Sample: |
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 dimer dimer, 32 kDa Human herpesvirus 8 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
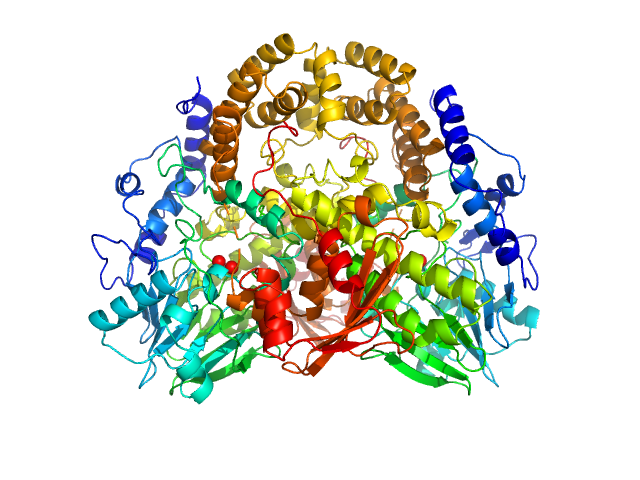
| Sample: |
Pseudomonas aeruginosa SDS hydrolase SdsA1 dimer, 145 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM HEPES, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2013 Jul 22
|
SdsA polymorph isolation and improvement of their crystal quality using nonconventional crystallization techniques
Journal of Applied Crystallography 48(5):1551-1559 (2015)
De la Mora E, Flores-Hernández E, Jakoncic J, Stojanoff V, Siliqi D, Sánchez-Puig N, Moreno A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
261 |
nm3 |
|
|
|
|
|
|
|
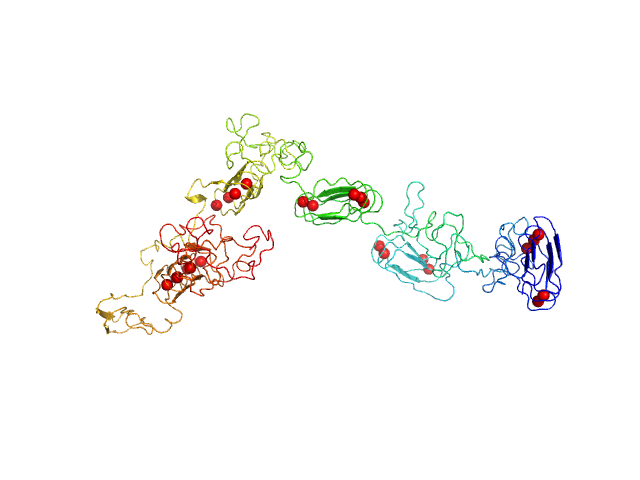
| Sample: |
RD domain of B. Pertussis Adenylate Cyclase Toxin (CyaA) monomer, 73 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM DTT, 4 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 31
|
Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion.
Sci Rep 5:14223 (2015)
O'Brien DP, Hernandez B, Durand D, Hourdel V, Sotomayor-Pérez AC, Vachette P, Ghomi M, Chamot-Rooke J, Ladant D, Brier S, Chenal A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
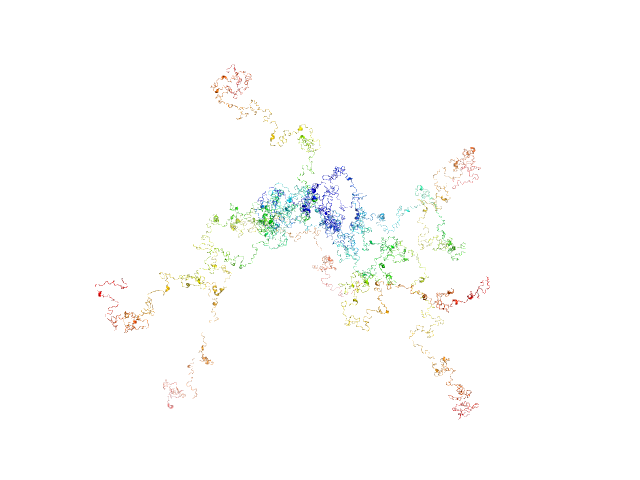
| Sample: |
RD domain of B. Pertussis Adenylate Cyclase Toxin (CyaA) monomer, 73 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 31
|
Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion.
Sci Rep 5:14223 (2015)
O'Brien DP, Hernandez B, Durand D, Hourdel V, Sotomayor-Pérez AC, Vachette P, Ghomi M, Chamot-Rooke J, Ladant D, Brier S, Chenal A
|
| RgGuinier |
8.3 |
nm |
| Dmax |
33.0 |
nm |
|
|