|
|
|
|
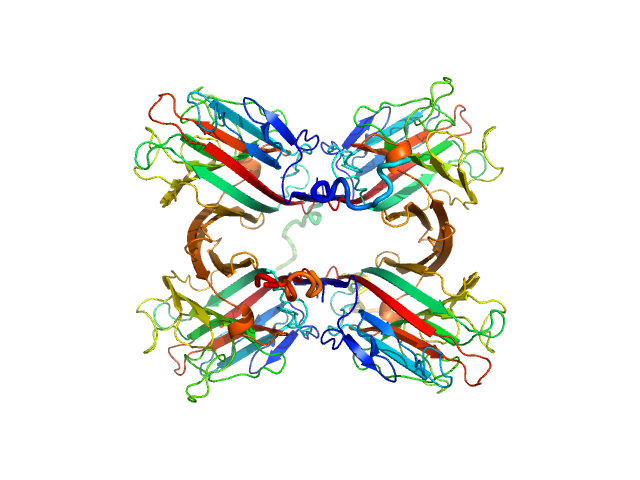
|
| Sample: |
Recombinant Tn antigen-binding lectin tetramer, 105 kDa Vatairea macrocarpa protein
|
| Buffer: |
100 mM sodium phosphate 150 mM NaCl 5% (v/v) glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 11
|
Structural characterization of a Vatairea macrocarpa lectin in complex with a tumor-associated antigen: A new tool for cancer research.
Int J Biochem Cell Biol 72:27-39 (2016)
Sousa BL, Silva-Filho JC, Kumar P, Graewert MA, Pereira RI, Cunha RMS, Nascimento KS, Bezerra GA, Delatorre P, Djinovic-Carugo K, Nagano CS, Gruber K, Cavada BS
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mouse Leucine-rich repeat transmembrane neuronal protein 2, cLRRTM2 (stability engineered construct) monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2015 Sep 27
|
Crystal Structure of an Engineered LRRTM2 Synaptic Adhesion Molecule and a Model for Neurexin Binding.
Biochemistry 55(6):914-26 (2016)
Paatero A, Rosti K, Shkumatov AV, Sele C, Brunello C, Kysenius K, Singha P, Jokinen V, Huttunen H, Kajander T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.1 |
nm |
|
|
|
|
|
|

|
| Sample: |
Mouse Leucine-rich repeat transmembrane neuronal protein 2, LRRTM2 dimer, 80 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2015 Jun 28
|
Crystal Structure of an Engineered LRRTM2 Synaptic Adhesion Molecule and a Model for Neurexin Binding.
Biochemistry 55(6):914-26 (2016)
Paatero A, Rosti K, Shkumatov AV, Sele C, Brunello C, Kysenius K, Singha P, Jokinen V, Huttunen H, Kajander T
|
| RgGuinier |
4.2 |
nm |
| Dmax |
21.6 |
nm |
|
|
|
|
|
|
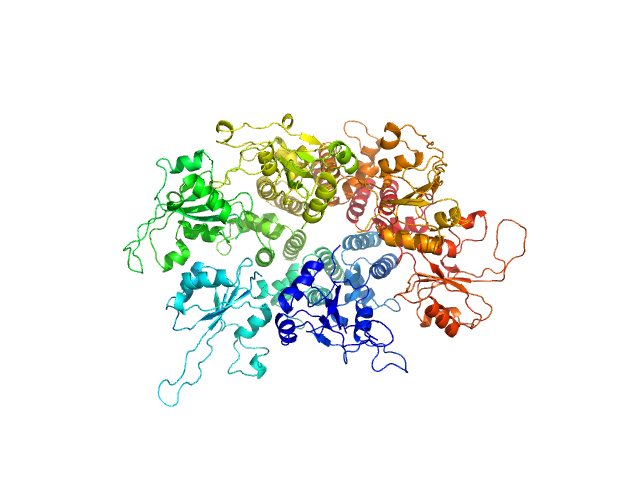
|
| Sample: |
Replication initiator protein of a promiscuous streptococcal plasmid pMV158. hexamer, Streptococcus sp. protein
|
| Buffer: |
10 mM TRIS 5 mM EDTA 1.0 M KCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Jun 16
|
Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep 6:20915 (2016)
Boer DR, Ruiz-Masó JA, Rueda M, Petoukhov MV, Machón C, Svergun DI, Orozco M, del Solar G, Coll M
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
282 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ankyrin repeat domains from Tankyrase 2 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human Arpin monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
2.6 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Zebrafish (Danio rerio) Arpin monomer, 25 kDa Danio rerio protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Zebrafish (Danio rerio) Arpin truncated C-terminal mutant (delta-C 16). monomer, 24 kDa Danio rerio protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Zebrafish arpin/human tankyrase 2 ankyrin repeat domain complex monomer, 43 kDa Danio rerio / … protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Middle East Respiratory Syndrome (MERS) coronavirus nucleocapsid N-Protein (N-terminal domain 1-164) monomer, 18 kDa Middle East respiratory … protein
|
| Buffer: |
10 mM HEPES 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Oct 5
|
Structural characterization of the N-terminal part of the MERS-CoV nucleocapsid by X-ray diffraction and small-angle X-ray scattering.
Acta Crystallogr D Struct Biol 72(Pt 2):192-202 (2016)
Papageorgiou N, Lichière J, Baklouti A, Ferron F, Sévajol M, Canard B, Coutard B
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|