|
|
|
|

|
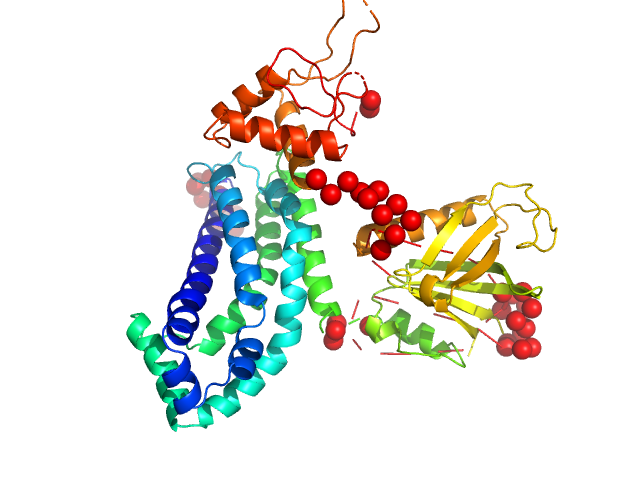
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 8 monomer, 127 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 18
|
Autoinhibition of ubiquitin-specific protease 8: insights into domain interactions and mechanisms of regulation
Journal of Biological Chemistry :107727 (2024)
Caba C, Black M, Liu Y, DaDalt A, Mallare J, Fan L, Harding R, Wang Y, Vacratsis P, Huang R, Zhuang Z, Tong Y
|
| RgGuinier |
8.4 |
nm |
| Dmax |
31.8 |
nm |
| VolumePorod |
327 |
nm3 |
|
|
|
|
|
|
|
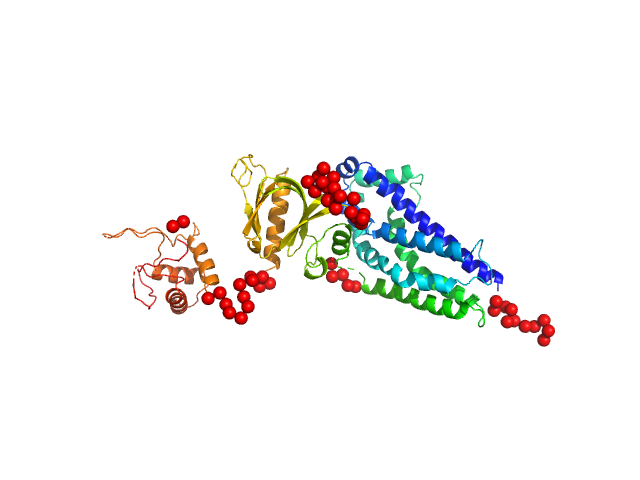
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
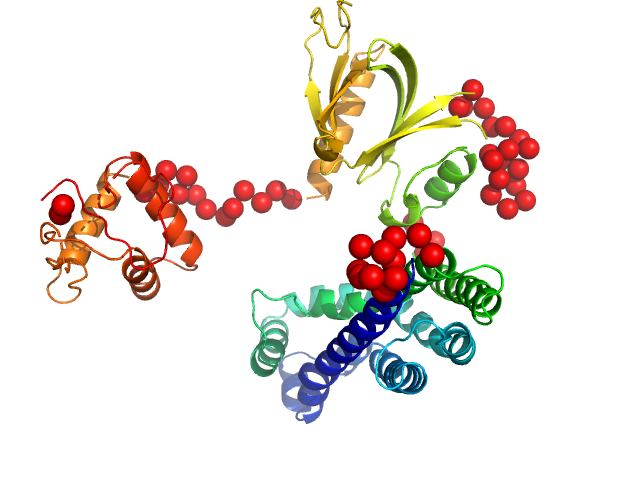
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (A170K) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 24
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
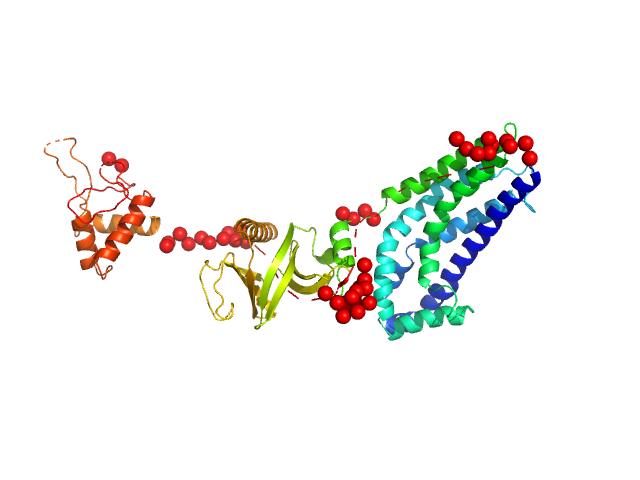
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (L177E) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (I409A) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
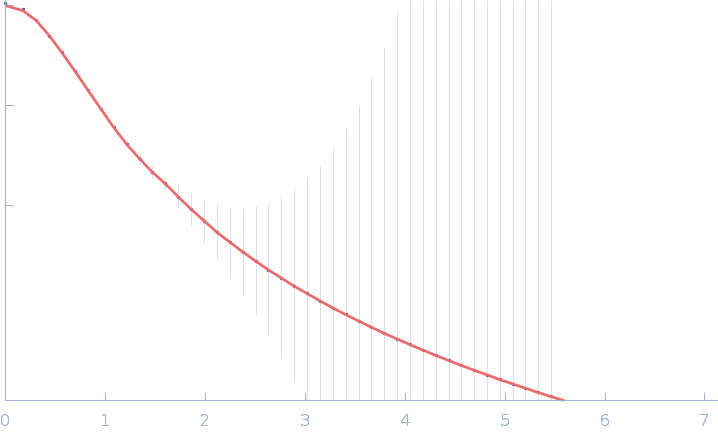
| Sample: |
Fas-activated serine/threonine kinase (amino acids 76-549) monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5% (v/v) Glycerol, 200 mM Ammonium Sulphate, 50 mM NaCl, 0.5 mM TCEP, 10 mM Glutamic acid, 10 mM Arginine;, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 31
|
Human FASTK protein
Daria Dawidziak
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
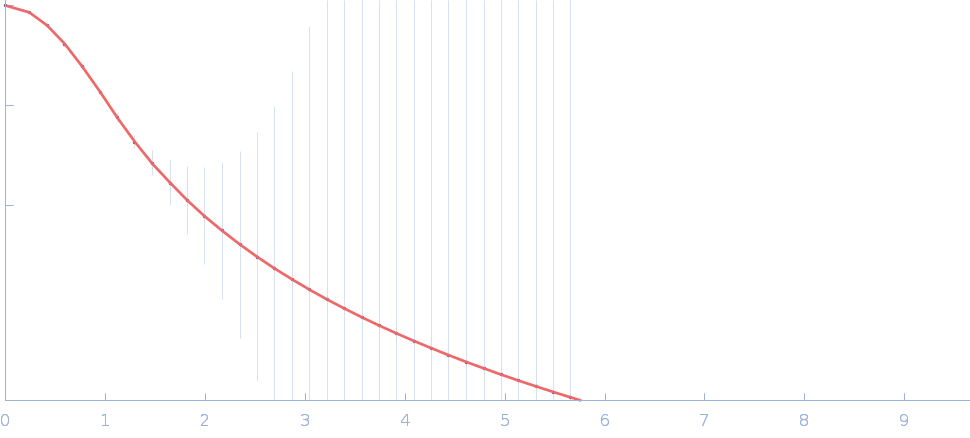
| Sample: |
Fas-activated serine/threonine kinase (amino acids 169-549) monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5% (v/v) Glycerol, 200 mM Ammonium Sulphate, 50 mM NaCl, 0.5 mM TCEP, 10 mM Glutamic acid, 10 mM Arginine;, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 31
|
Human FASTK protein
Daria Dawidziak
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1 (C43S/C175S) monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 7
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1 (C43S/C175S) monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Sep 27
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1 (C43S/C175S) monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 1
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
|
|