|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Jun 8
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Jun 8
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Jun 8
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Jun 8
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
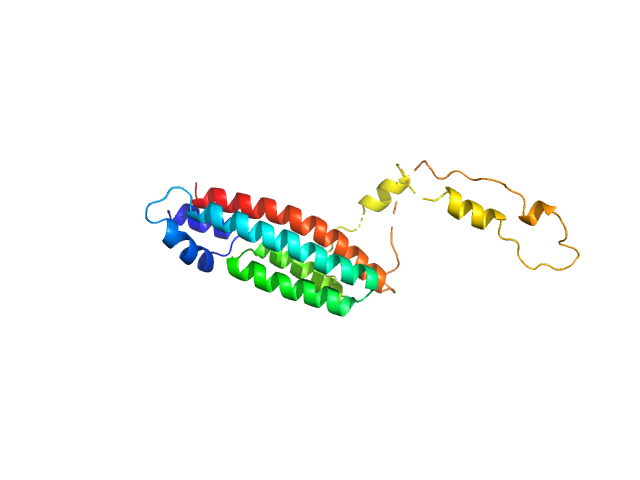
| Sample: |
Type 2 DNA topoisomerase 6 subunit B-like monomer, 61 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 500 mM KCl, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 12
|
The TOPOVIBL meiotic DSB formation protein: new insights from its biochemical and structural characterization
Nucleic Acids Research (2024)
Diagouraga B, Tambones I, Carivenc C, Bechara C, Nadal M, de Massy B, le Maire A, Robert T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphorelay intermediate protein YPD1 monomer, 19 kDa Candida albicans (strain … protein
|
| Buffer: |
50 mM Tris-HCl pH 8, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Oct 10
|
Structural and functional insights underlying recognition of histidine phosphotransfer protein in fungal phosphorelay systems
Communications Biology 7(1) (2024)
Paredes-Martínez F, Eixerés L, Zamora-Caballero S, Casino P
|
| RgGuinier |
2.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
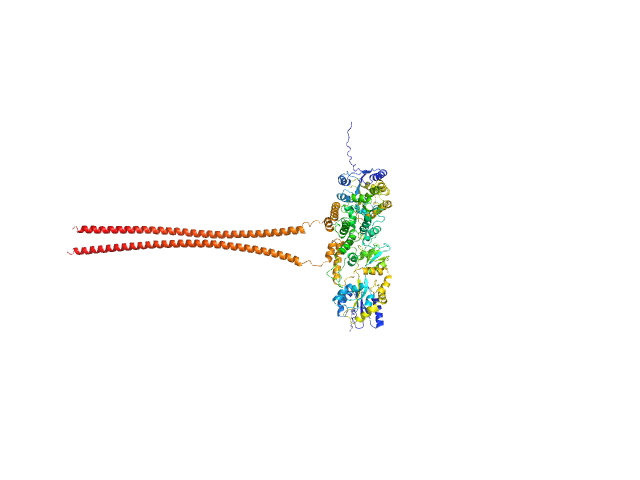
| Sample: |
Septin-10 dimer, 110 kDa Schistosoma mansoni protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 17
|
Novel lipid-interaction motifs within the C-terminal domain of Septin10 from Schistosoma mansoni
Biochimica et Biophysica Acta (BBA) - Biomembranes :184371 (2024)
Cavini I, Fontes M, Zeraik A, Lopes J, Araújo A
|
| RgGuinier |
5.7 |
nm |
| Dmax |
27.4 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
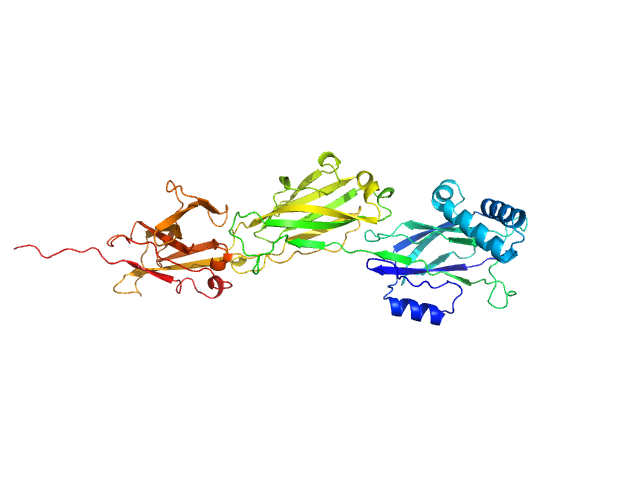
| Sample: |
LPXTG-motif cell wall anchor domain protein monomer, 49 kDa Ligilactobacillus ruminis ATCC … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 1
|
The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis
Acta Crystallographica Section D Structural Biology 80(7):474-492 (2024)
Prajapati A, Palva A, von Ossowski I, Krishnan V
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
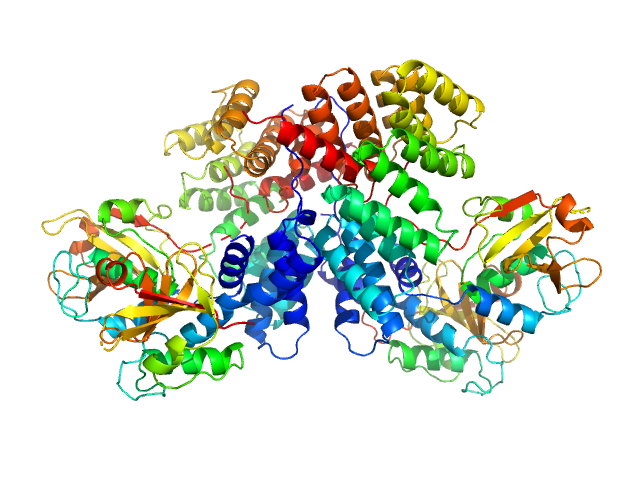
| Sample: |
Lipoprotein NlpI dimer, 63 kDa Escherichia coli (strain … protein
Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase tetramer, 73 kDa Escherichia coli (strain … protein
|
| Buffer: |
0.2 M Na/K phosphate pH 7.0, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2023 Sep 7
|
Structural basis for recruitment of peptidoglycan endopeptidase MepS by lipoprotein NlpI.
Nat Commun 15(1):5461 (2024)
Wang S, Huang CH, Lin TS, Yeh YQ, Fan YS, Wang SW, Tseng HC, Huang SJ, Chang YY, Jeng US, Chang CI, Tzeng SR
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Pigeon iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Pigeon iron-sulfur cluster assembly 1 homolog (clISCA1) 29.3 mg/ml
Shigeki Arai
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
130 |
nm3 |
|
|