|
|
|
|

|
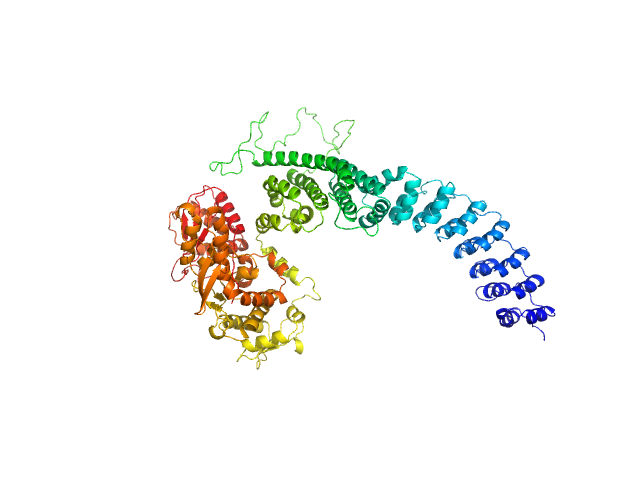
| Sample: |
E3 ubiquitin-protein ligase HACE1 dimer, 205 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)
Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens T, Bohnsack K, Urlaub H, Steinchen W, Dienemann C, Lorenz S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
379 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase HACE1 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)
Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens T, Bohnsack K, Urlaub H, Steinchen W, Dienemann C, Lorenz S
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
|
|
|
|
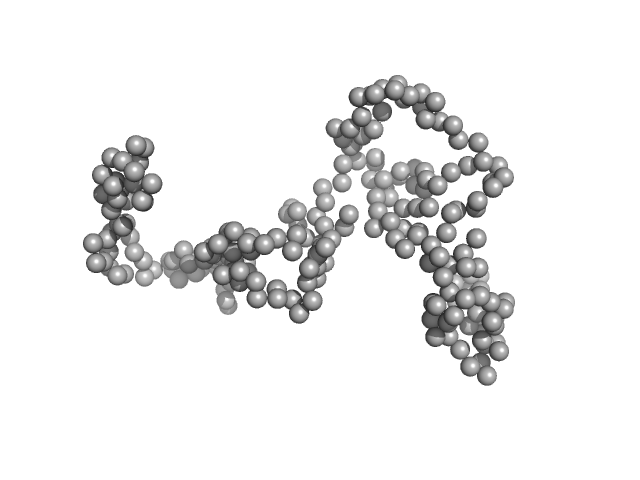
|
| Sample: |
DTMP kinase dimer, 57 kDa Brugia malayi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2018 Oct 7
|
Crystal Structure of the Brugia malayi Thymidylate Kinase-dTMP Complex and Small Angle X-ray Scattering Experiments Identifies Changes in the Dimeric Association Compared to the Human Homolog
Crystallography Reports 68(7):1150-1158 (2024)
Vishwakarma J, Sharma V, Kumar S, Ramachandran R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
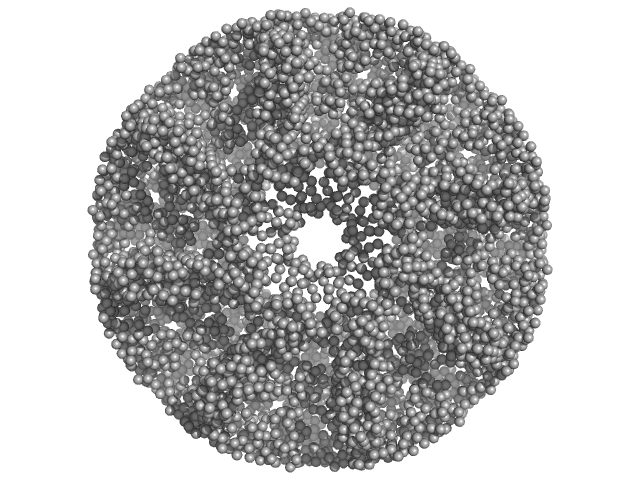
| Sample: |
Carboxypeptidase-related protein 16-mer, 903 kDa Deinococcus radiodurans (strain … protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Dec 25
|
Novel oligomeric assembly of S10-carboxypeptidase from Deinococcus radiodurans
Dr. Rahul Singh
|
| RgGuinier |
7.4 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
1180 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 27
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
705 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 27
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
704 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 27
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
702 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 29
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
699 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 30
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
706 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 30
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
706 |
nm3 |
|
|