|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 441-543) monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.00, 500 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|

|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
Fragment antigen-binding region Fab10C12 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 3
|
Inhibition of aggregation and biofilm formation by Uropathogenic Escherichia coli
Andrew Whitten
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translocated intimin receptor Tir dimer, 50 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 18
|
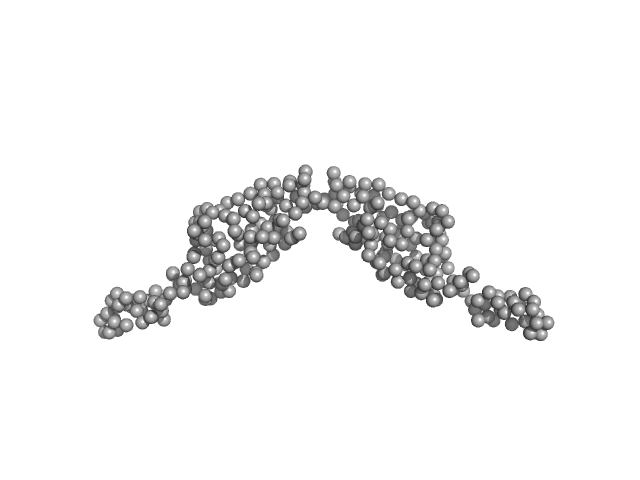
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translocated intimin receptor Tir dimer, 32 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translocated intimin receptor Tir monomer, 18 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
20 mM Sodium Phosphate, 150 mM NaCl, 1 mM EDTA, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 15
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translocated intimin receptor Tir monomer, 12 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Nov 21
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
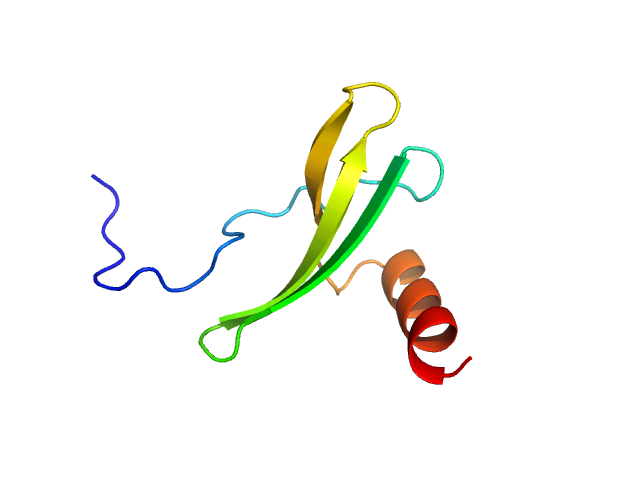
| Sample: |
NHLP leader peptide family natural product (I56V) monomer, 9 kDa Methylovulum psychrotolerans protein
|
| Buffer: |
20 mM sodium phosphate (pH 7.5), 100 mM NaCl and 48 µM FAD, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Pennsylvania State University on 2023 Jul 28
|
Disordered regions in proteusin peptides guide post-translational modification by a flavin-dependent RiPP brominase.
Nat Commun 15(1):1265 (2024)
Nguyen NA, Vidya FNU, Yennawar NH, Wu H, McShan AC, Agarwal V
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stromal cell-derived factor 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
High mobility group protein B1 (D189E, E202D, E215D) monomer, 25 kDa Rattus norvegicus protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stromal cell-derived factor 1 monomer, 9 kDa Homo sapiens protein
High mobility group protein B1 (D189E, E202D, E215D) monomer, 25 kDa Rattus norvegicus protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
72 |
nm3 |
|
|