|
|
|
|
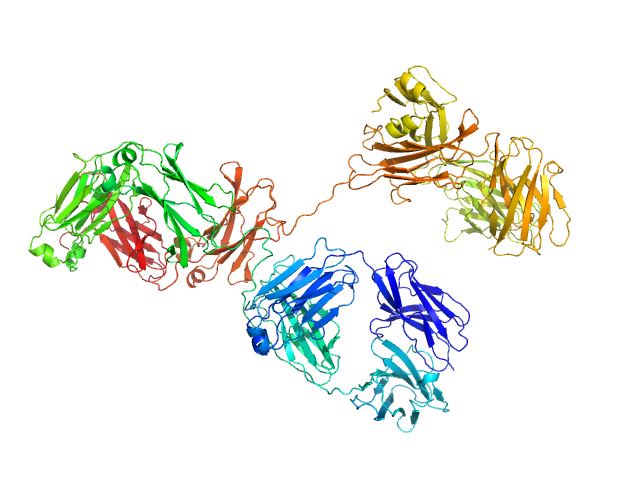
|
| Sample: |
Immunoglobulin G subclass 4 monomer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Using atomistic solution scattering modelling to elucidate the role of the Fc glycans in human IgG4.
PLoS One 19(4):e0300964 (2024)
Spiteri VA, Doutch J, Rambo RP, Bhatt JS, Gor J, Dalby PA, Perkins SJ
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
|
|
|
|
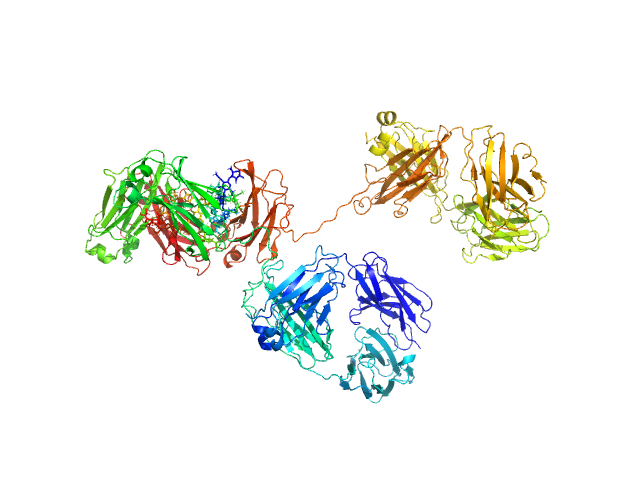
|
| Sample: |
Immunoglobulin G subclass 4 monomer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Using atomistic solution scattering modelling to elucidate the role of the Fc glycans in human IgG4.
PLoS One 19(4):e0300964 (2024)
Spiteri VA, Doutch J, Rambo RP, Bhatt JS, Gor J, Dalby PA, Perkins SJ
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
|
|
|
|
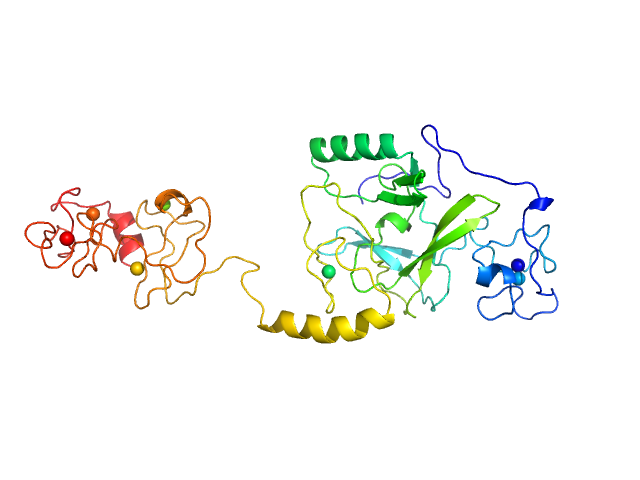
|
| Sample: |
Histone-lysine N-methyltransferase NSD3 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
0.5 M NaCl, 20 mM Tris-HCl, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Nov 25
|
Structural insights into the C-terminus of the histone-lysine N-methyltransferase NSD3 by small-angle X-ray scattering.
Front Mol Biosci 11:1191246 (2024)
Belviso BD, Shen Y, Carrozzini B, Morishita M, di Luccio E, Caliandro R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
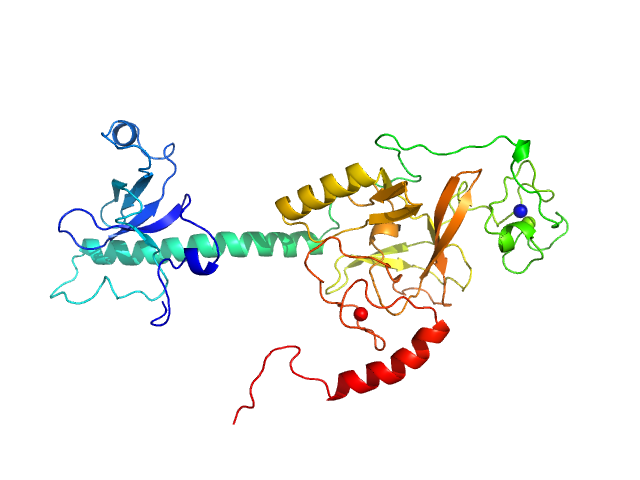
|
| Sample: |
Histone-lysine N-methyltransferase NSD3 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
0.5 M NaCl, 20 mM Tris-HCl, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Nov 25
|
Structural insights into the C-terminus of the histone-lysine N-methyltransferase NSD3 by small-angle X-ray scattering.
Front Mol Biosci 11:1191246 (2024)
Belviso BD, Shen Y, Carrozzini B, Morishita M, di Luccio E, Caliandro R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
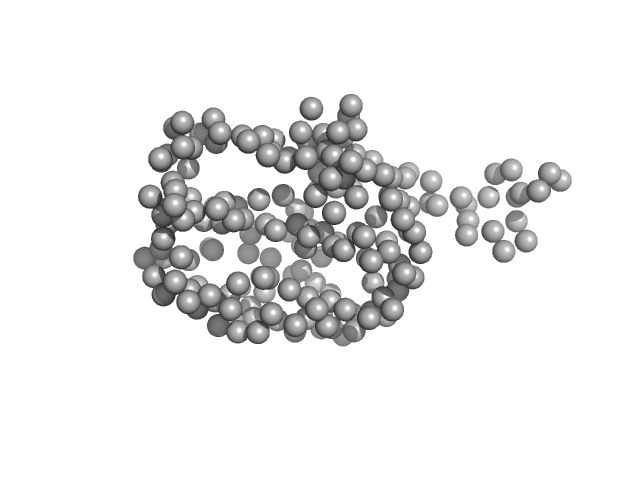
|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Biofilm regulatory protein monomer, 34 kDa Streptococcus dysgalactiae subsp. … protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 1
|
Structural insights of an LCP protein-LytR-from Streptococcus dysgalactiae subs. dysgalactiae through biophysical and in silico methods.
Front Chem 12:1379914 (2024)
Paquete-Ferreira J, Freire F, Fernandes HS, Muthukumaran J, Ramos J, Bryton J, Panjkovich A, Svergun D, Santos MFA, Correia MAS, Fernandes AR, Romão MJ, Sousa SF, Santos-Silva T
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
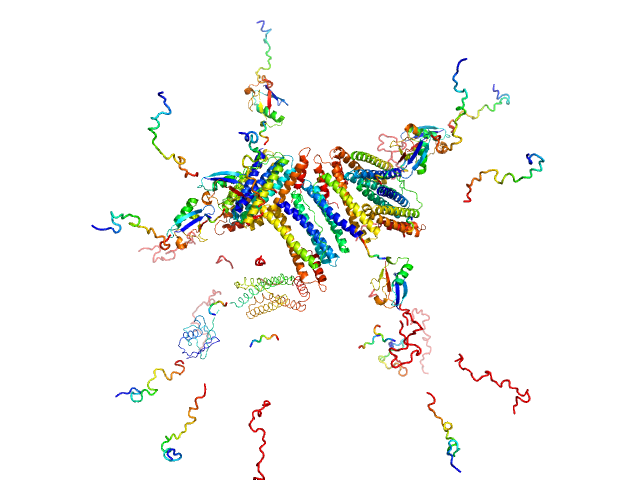
| Sample: |
Biofilm regulatory protein monomer, 34 kDa Streptococcus dysgalactiae subsp. … protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 1
|
Structural insights of an LCP protein-LytR-from Streptococcus dysgalactiae subs. dysgalactiae through biophysical and in silico methods.
Front Chem 12:1379914 (2024)
Paquete-Ferreira J, Freire F, Fernandes HS, Muthukumaran J, Ramos J, Bryton J, Panjkovich A, Svergun D, Santos MFA, Correia MAS, Fernandes AR, Romão MJ, Sousa SF, Santos-Silva T
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion) 24-mer, 755 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Jul 30
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
7.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
2010 |
nm3 |
|
|