|
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK monomer, 77 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
4.1 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK (A384P, S386P, T387P, A388P, L390F) monomer, 77 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
3.9 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK (SH3-SH2-kinase domains) monomer, 52 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
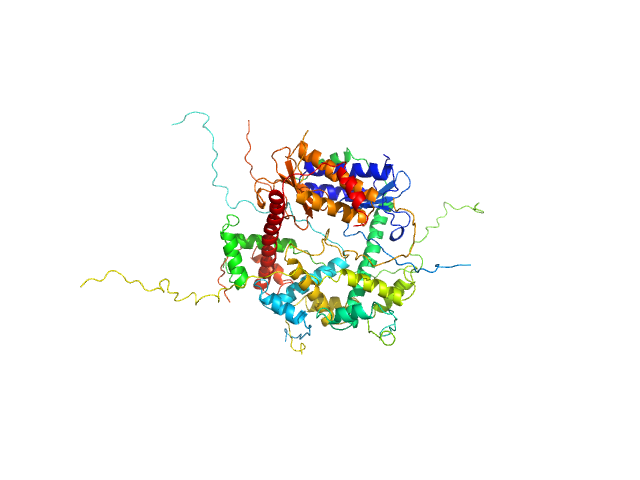
| Sample: |
Tyrosine-protein kinase BTK (SH3-SH2-kinase domain A384P, S386P, T387P, A388P, L390F) monomer, 53 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
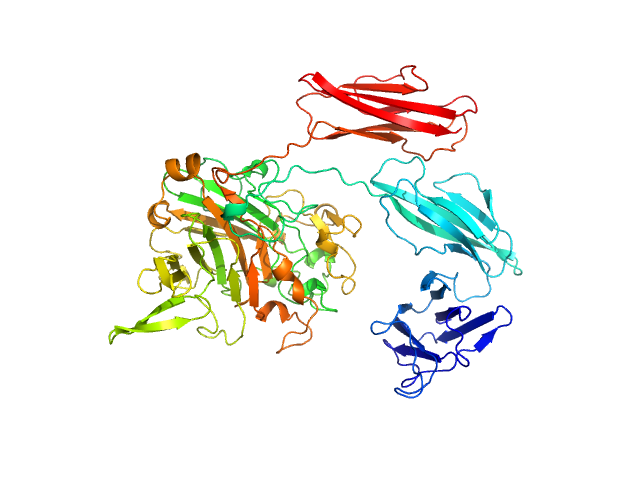
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 6
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase isozyme L1 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.03% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 Sep 2
|
Altered protein dynamics and a more reactive catalytic cysteine in a neurodegeneration-associated UCHL1 mutant.
J Mol Biol :168438 (2024)
Kenny S, Lai CH, Chiang TS, Brown K, Hewitt CS, Krabill AD, Chang HT, Wang YS, Flaherty DP, Danny Hsu ST, Das C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase isozyme L1 (R178Q) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.03% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 Sep 2
|
Altered protein dynamics and a more reactive catalytic cysteine in a neurodegeneration-associated UCHL1 mutant.
J Mol Biol :168438 (2024)
Kenny S, Lai CH, Chiang TS, Brown K, Hewitt CS, Krabill AD, Chang HT, Wang YS, Flaherty DP, Danny Hsu ST, Das C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dockerin type I repeat monomer, 69 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
The Ruminococcus bromii amylosome protein Sas6 binds single and double helical α-glucan structures in starch.
Nat Struct Mol Biol (2024)
Photenhauer AL, Villafuerte-Vega RC, Cerqueira FM, Armbruster KM, Mareček F, Chen T, Wawrzak Z, Hopkins JB, Vander Kooi CW, Janeček Š, Ruotolo BT, Koropatkin NM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform monomer, 65 kDa Homo sapiens protein
Isoform Delta-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (R160H) monomer, 70 kDa Homo sapiens protein
Isoform 1 of Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 50 µM MnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 20
|
B56δ long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc Natl Acad Sci U S A 121(1):e2310727120 (2024)
Wu CG, Balakrishnan VK, Merrill RA, Parihar PS, Konovolov K, Chen YC, Xu Z, Wei H, Sundaresan R, Cui Q, Wadzinski BE, Swingle MR, Musiyenko A, Chung WK, Honkanen RE, Suzuki A, Huang X, Strack S, Xing Y
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
266 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform monomer, 65 kDa Homo sapiens protein
Isoform 1 of Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform monomer, 36 kDa Homo sapiens protein
Isoform Delta-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (R160H, E198K) monomer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 50 µM MnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 20
|
B56δ long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc Natl Acad Sci U S A 121(1):e2310727120 (2024)
Wu CG, Balakrishnan VK, Merrill RA, Parihar PS, Konovolov K, Chen YC, Xu Z, Wei H, Sundaresan R, Cui Q, Wadzinski BE, Swingle MR, Musiyenko A, Chung WK, Honkanen RE, Suzuki A, Huang X, Strack S, Xing Y
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
305 |
nm3 |
|
|