|
|
|
|
|
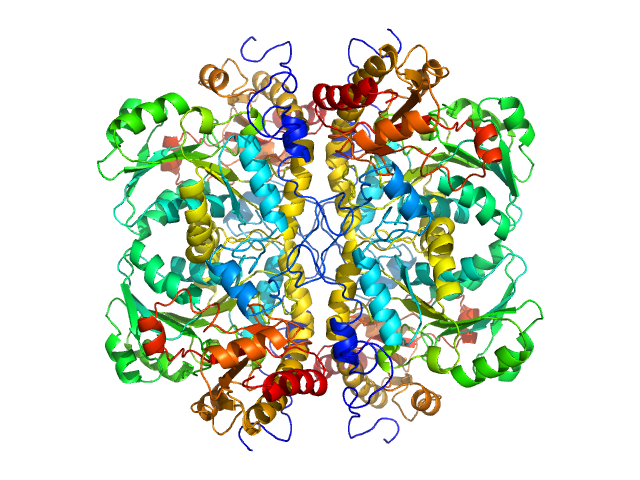
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
11.0 |
nm |
| Dmax |
70.0 |
nm |
| VolumePorod |
1900 |
nm3 |
|
|
|
|
|
|
|
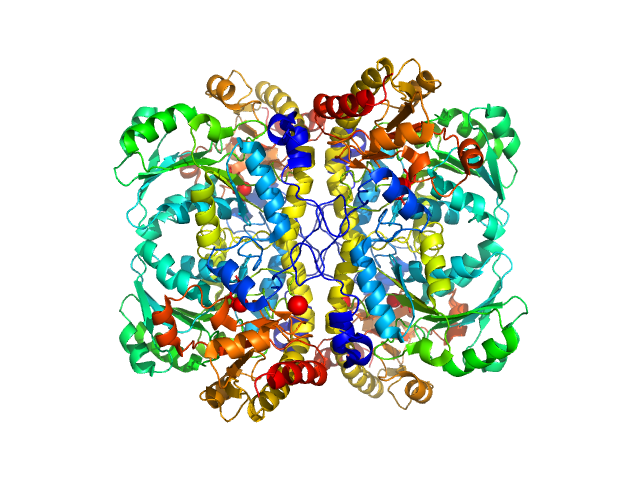
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
11.3 |
nm |
| Dmax |
66.0 |
nm |
| VolumePorod |
1820 |
nm3 |
|
|
|
|
|
|
|
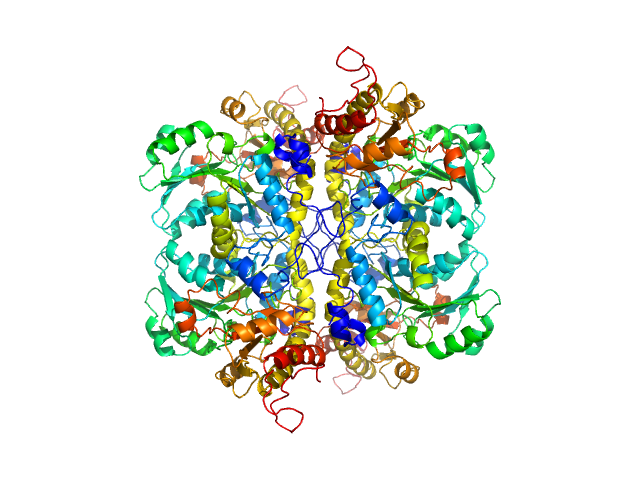
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
12.5 |
nm |
| Dmax |
65.0 |
nm |
| VolumePorod |
2044 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
14.2 |
nm |
| Dmax |
65.0 |
nm |
| VolumePorod |
2190 |
nm3 |
|
|
|
|
|
|
|
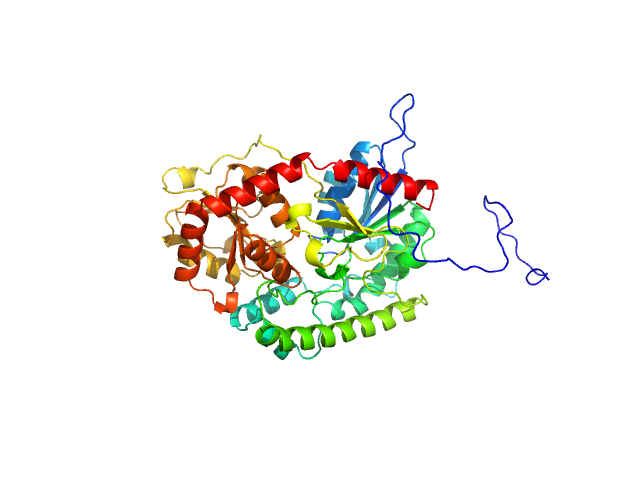
| Sample: |
L-methionine gamma-lyase tetramer, 181 kDa Clostridium tetani protein
|
| Buffer: |
PBS-D2O: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (D2O buffer), pH: 7.4 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 May 19
|
Methionine gamma lyase fused with S3 domain VGF forms octamers and adheres to tumor cells via binding EGFR
Biochemical and Biophysical Research Communications :149319 (2023)
Bondarev N, Bagaeva D, Bazhenov S, Buben M, Bulushova N, Ryzhykau Y, Okhrimenko I, Zagryadskaya Y, Maslov I, Anisimova N, Sokolova D, Kuklin A, Pokrovsky V, Manukhov I
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
232 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
L-methionine gamma-lyase (K272S) tetramer, 174 kDa Clostridium sporogenes protein
|
| Buffer: |
PBS-D2O: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (D2O buffer), pH: 7.4 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 May 19
|
Methionine gamma lyase fused with S3 domain VGF forms octamers and adheres to tumor cells via binding EGFR
Biochemical and Biophysical Research Communications :149319 (2023)
Bondarev N, Bagaeva D, Bazhenov S, Buben M, Bulushova N, Ryzhykau Y, Okhrimenko I, Zagryadskaya Y, Maslov I, Anisimova N, Sokolova D, Kuklin A, Pokrovsky V, Manukhov I
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
L-methionine gamma-lyase from Clostridium sporogenes fused with VGF S3 domain tetramer, 183 kDa Clostridium sporogenes protein
|
| Buffer: |
PBS-D2O: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (D2O buffer), pH: 7.4 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 May 19
|
Methionine gamma lyase fused with S3 domain VGF forms octamers and adheres to tumor cells via binding EGFR
Biochemical and Biophysical Research Communications :149319 (2023)
Bondarev N, Bagaeva D, Bazhenov S, Buben M, Bulushova N, Ryzhykau Y, Okhrimenko I, Zagryadskaya Y, Maslov I, Anisimova N, Sokolova D, Kuklin A, Pokrovsky V, Manukhov I
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.3 |
nm |
| VolumePorod |
303 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
UDP-glycosyltransferase 202A2 monomer, 53 kDa Tetranychus urticae protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 17
|
Structural and functional studies reveal the molecular basis of substrate promiscuity of a glycosyltransferase originating from a major agricultural pest
Journal of Biological Chemistry :105421 (2023)
Arriaza R, Abiskaroon B, Patel M, Daneshian L, Kluza A, Snoeck S, Watkins M, Hopkins J, Van Leeuwen T, Grbic M, Grbic V, Borowski T, Chruszcz M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
20 kDa accessory protein dimer, 42 kDa Bacillus thuringiensis serovar … protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2023 Aug 15
|
20-kDa accessory protein (P20) from Bacillus thuringiensis subsp. israelensis ISPC-12: Purification, characterization, solution scattering and structural analysis
International Journal of Biological Macromolecules :127985 (2023)
Kinkar O, Singh R, Prashar A, Kumar A, Hire R, Makde R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 0.1 mM CaCl2, 1 mM NaN3, 0.2 mM ATP, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 1
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|