|
|
|
|

|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 0.1 mM CaCl2, 1 mM NaN3, 1.0 mM ATP, 50 mM KCl, 2 mM MgCl2, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 1
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
15.7 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
4710 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDTZ4_fit1_model1.png)
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 2.0 mM EDTA, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Nov 16
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
549 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDT25_fit1_model1.png)
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris, 0.1 mM CaCl2, 1 mM NaN3, 0.2 mM ATP, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 5
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Heat-labile enterotoxin B chain monomer, 36 kDa Clostridium perfringens protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 18
|
Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin
Toxins 15(11):637 (2023)
Ogbu C, Kapoor S, Vecchio A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Heat-labile enterotoxin B chain monomer, 33 kDa Clostridium perfringens protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 18
|
Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin
Toxins 15(11):637 (2023)
Ogbu C, Kapoor S, Vecchio A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Heat shock factor 2-binding protein tetramer, 150 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 250 mM NaCl, and 5 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Oct 2
|
BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv 9(43):eadi7352 (2023)
Ghouil R, Miron S, Sato K, Ristic D, van Rossum-Fikkert SE, Legrand P, Ouldali M, Winter JM, Ropars V, David G, Arteni AA, Wyman C, Knipscheer P, Kanaar R, Zelensky AN, Zinn-Justin S
|
| RgGuinier |
8.5 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
364 |
nm3 |
|
|
|
|
|
|
|
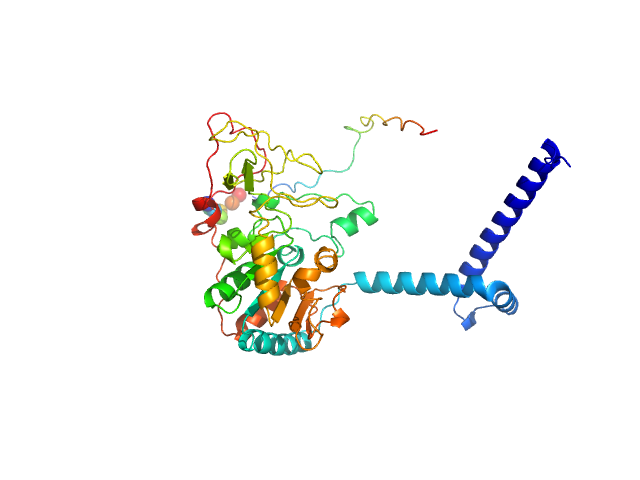
| Sample: |
Isoform 1 of NAD-dependent protein deacetylase sirtuin-7 monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 16
|
Substrates and Cyclic Peptide Inhibitors of the Oligonucleotide-Activated Sirtuin 7.
Angew Chem Int Ed Engl :e202314597 (2023)
Bolding JE, Nielsen AL, Jensen I, Hansen TN, Ryberg LA, Jameson ST, Harris P, Peters GHJ, Denu JM, Rogers JM, Olsen CA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 14
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 15
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|
|
|
|
|
|
| Sample: |
Ferritin light chain 24-mer, 479 kDa Equus caballus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 14
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|