|
|
|
|
|
| Sample: |
Human Latent Transforming Growth Factor beta 1 dimer, 86 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Oct 4
|
Structural consequences of transforming growth factor beta-1 activation from near-therapeutic X-ray doses.
J Synchrotron Radiat 26(Pt 4):967-979 (2019)
Stachowski T, Grant TD, Snell EH
|
| RgGuinier |
3.8 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
200 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Latency Associated Peptide dimer, 58 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Oct 4
|
Structural consequences of transforming growth factor beta-1 activation from near-therapeutic X-ray doses.
J Synchrotron Radiat 26(Pt 4):967-979 (2019)
Stachowski T, Grant TD, Snell EH
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
179 |
nm3 |
|
|
|
|
|
|
|
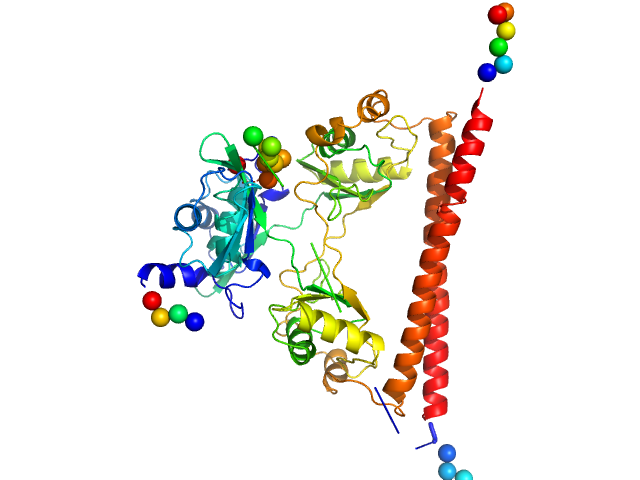
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 250 mM NaCl, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 19
|
A new crystal structure and small-angle X-ray scattering analysis of the homodimer of human SFPQ.
Acta Crystallogr F Struct Biol Commun 75(Pt 6):439-449 (2019)
Hewage TW, Caria S, Lee M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
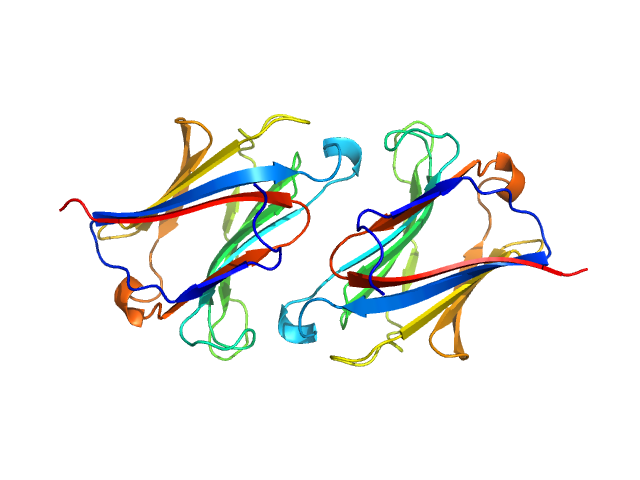
| Sample: |
Galectin-10 Tyr69Glu dimer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes 150 NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 4
|
Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science 364(6442) (2019)
Persson EK, Verstraete K, Heyndrickx I, Gevaert E, Aegerter H, Percier JM, Deswarte K, Verschueren KHG, Dansercoer A, Gras D, Chanez P, Bachert C, Gonçalves A, Van Gorp H, De Haard H, Blanchetot C, Saunders M, Hammad H, Savvides SN, Lambrecht BN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jul 23
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jul 23
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) mutant I396E monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 9
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
5.3 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jul 23
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
131 |
nm3 |
|
|
|
|
|
|
|
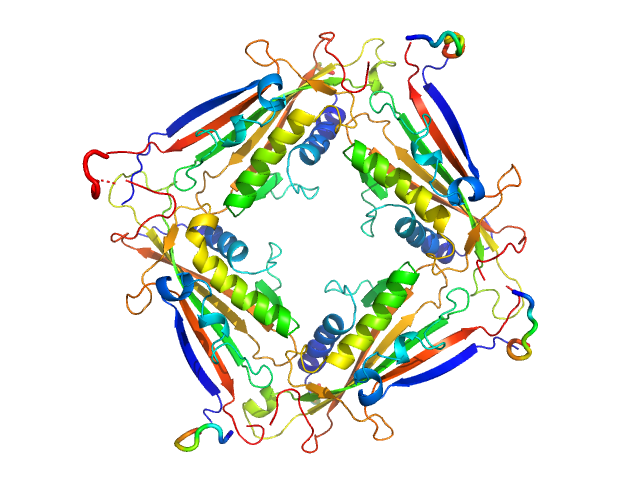
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
|
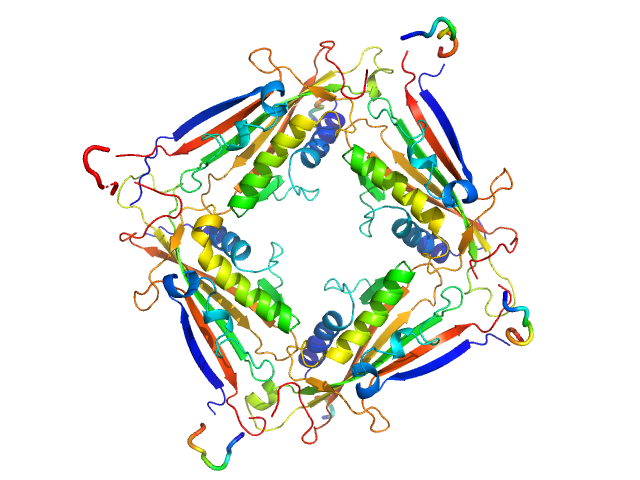
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|