|
|
|
|
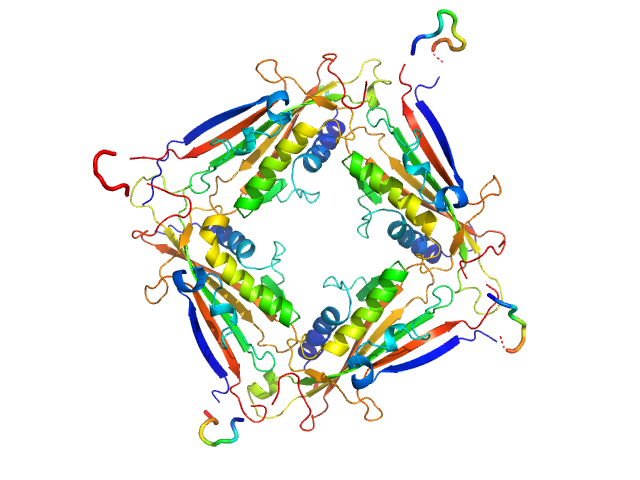
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 6.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
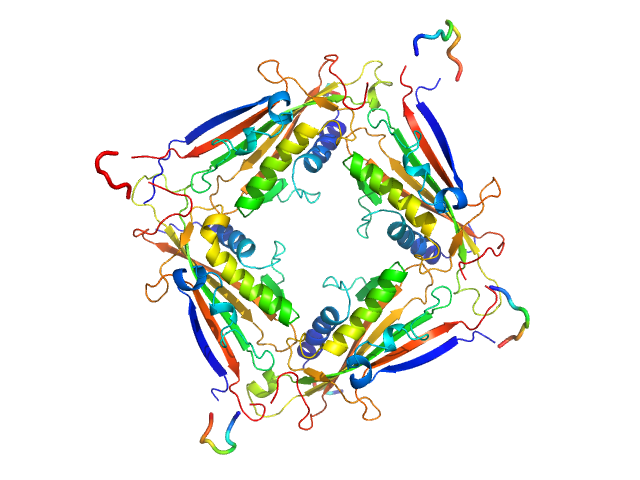
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 6.5 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
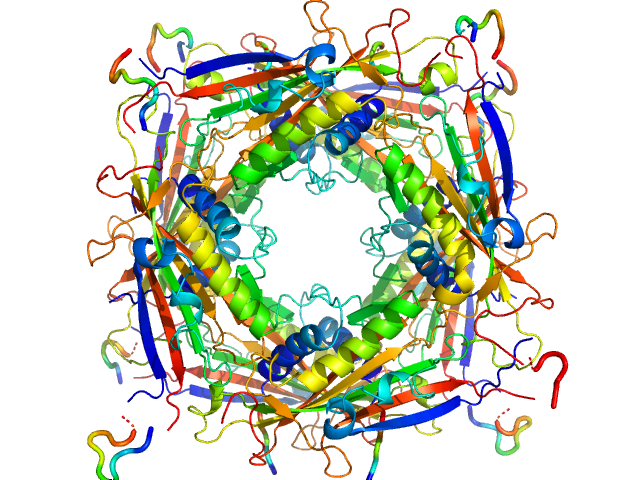
|
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
285 |
nm3 |
|
|
|
|
|
|
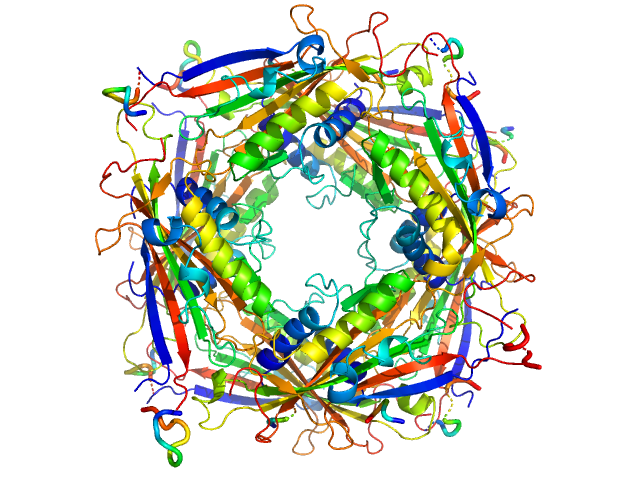
|
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E2 tetramer, 139 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
The molecular basis for Apolipoprotein E4 as the major risk factor for late onset Alzheimer's disease.
J Mol Biol (2019)
Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, Serpell L
|
| RgGuinier |
5.6 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E3 tetramer, 139 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
The molecular basis for Apolipoprotein E4 as the major risk factor for late onset Alzheimer's disease.
J Mol Biol (2019)
Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, Serpell L
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
410 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein E4 tetramer, 139 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
The molecular basis for Apolipoprotein E4 as the major risk factor for late onset Alzheimer's disease.
J Mol Biol (2019)
Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, Serpell L
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.6 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysyne-specific Demethylase LSD2 monomer, 89 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 11
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NPAC dehydrogenase domain tetramer, 125 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 26
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NPAC linker+DH (delta-205) tetramer, 150 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 26
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
240 |
nm3 |
|
|