|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
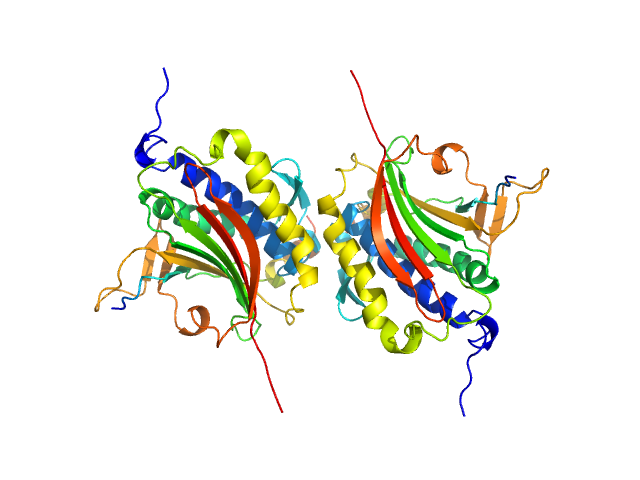
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
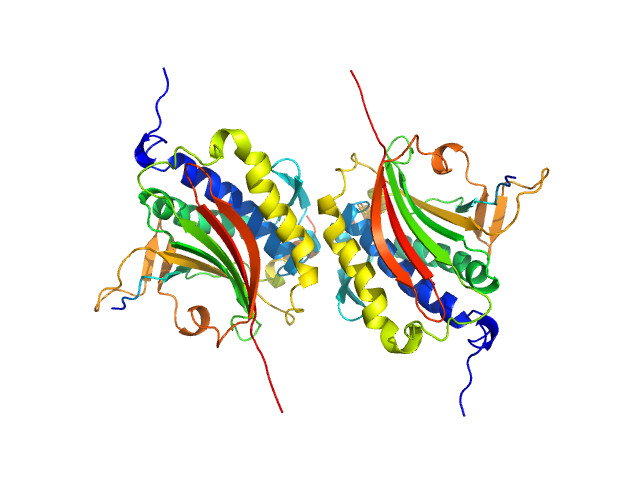
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
|
|
|
|
|
|

|
| Sample: |
Procollagen lysyl hydroxylase LH3 dimer, 180 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 28
|
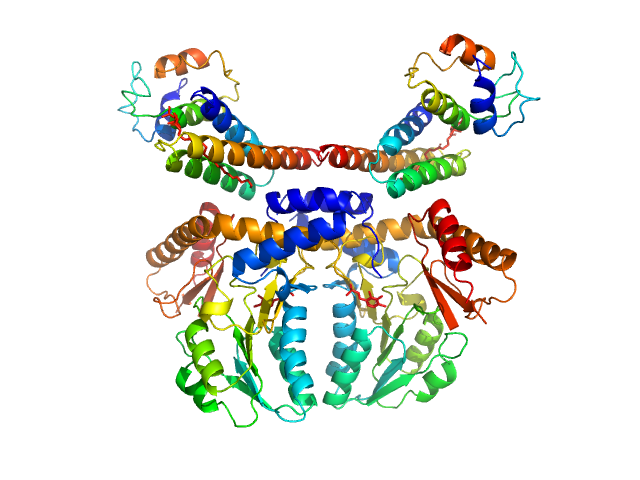
Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun 9(1):3163 (2018)
Scietti L, Chiapparino A, De Giorgi F, Fumagalli M, Khoriauli L, Nergadze S, Basu S, Olieric V, Cucca L, Banushi B, Profumo A, Giulotto E, Gissen P, Forneris F
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
268 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
204 |
nm3 |
|
|